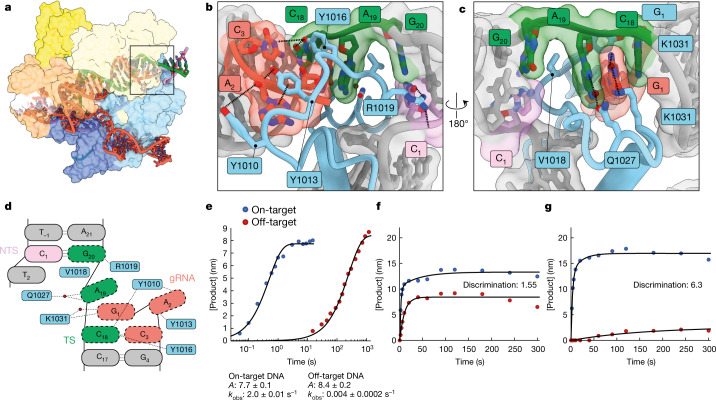
Fig. 4. Stabilization of distorted 18–20 MM by the RuvC domain and improved fidelity of SuperFi-Cas9.
a, Overall structure of the 18–20 MM active conformation viewed from the back. b, c, Magnified views of Cas9 interacting with the distal end of the duplex. Flipped gRNA base position 2 is accommodated by stacking interactions and hydrogen bonding with RuvC tyrosine side-chains, whereas a network of interactions (including a water-mediated hydrogen bond) stabilizes the stretched target strand configuration, which allows TS(20) to resume base-pairing with the NTS. d, Schematic of distorted PAM-distal gRNA–TS duplex. Red circles correspond to water molecules. e, Kinetics of on-target and off-target (18–20 MM) Mg2+-initiated cleavage by the 7-D Cas9 mutant (SuperFi-Cas9). f, g, Cleavage competition assay for wild-type Cas9 (f) and SuperFi-Cas9 (g). 25 nM of either Cas9 variant was mixed with 50 nM of each substrate and the cleaved DNA product was monitored. Discrimination in favour of the on-target DNA is defined by the ratio of amplitudes for on-target and off-target product formed.