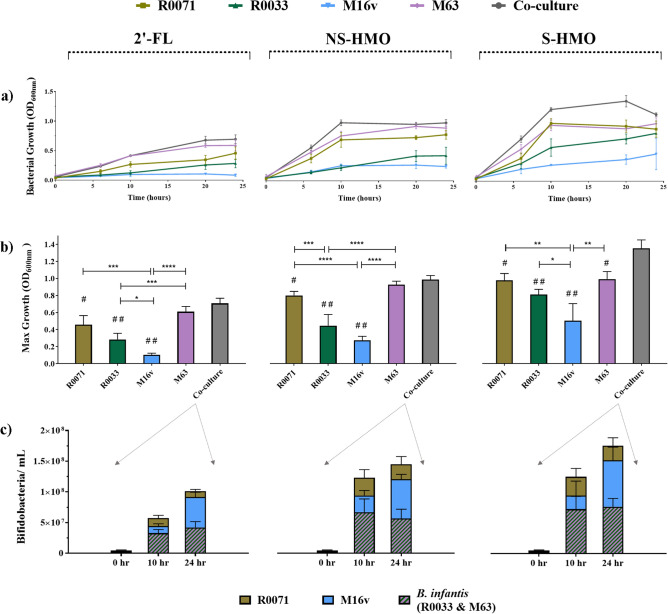
Figure 1.
Growth profiles of bifidobacterial strains as mono-cultures and as a four strain co-culture with the following HMO as the sole carbon source: 2’-FL [1.2 g/L], model of NS-HMO [3.8 g/L] and model of S-HMO [5 g/L]. (a) Growth of strains in mono- and co-culture at 0 h, 6 h, 10 h, 20 h and 24 h. (b) Maximum levels of growth reached by the strains in mono- and co-culture (c) Growth (bifidobacteria/mL) of subspecies within the four strain co-culture at 0 h, 10 h and 24 h. Strains within co-culture were quantified using qPCR with primers targeting the 16 s gene region of each subspecies. OD600nm = Optical density at 600 nm. R0071 = Bifidobacterium bifidum R0071, R0033 = Bifidobacterium infantis R0033, M-16V = Bifidobacterium breve M-16V, M-63 = Bifidobacterium infantis M-63. Analysis was calculated using technical triplicate data from biological triplicate experiments and data are means + /−SD. Univariate analysis of variance (ANOVA) and post-hoc Tukey tests were performed to determine the significant differences between the groups (# = p < 0.05, ## = p < 0.001 vs. co-culture) (* = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001 between other strain combinations).