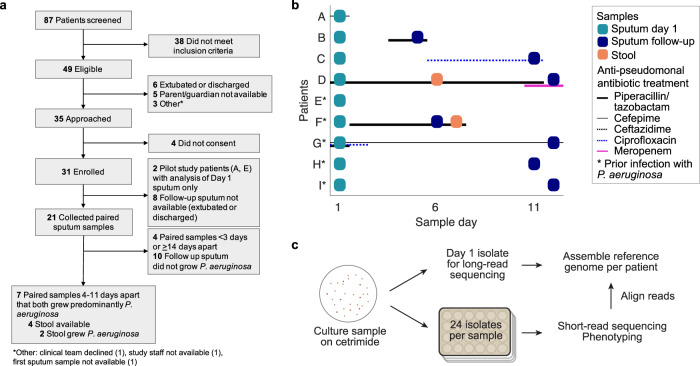
Fig. 1. Prospective study of P. aeruginosa populations from mechanically ventilated patients during acute respiratory tract infection.
a Prospective study design describing the enrollment strategy of mechanically ventilated patients in the ICU. Of 87 patients screened, 49 eligible patients were identified, from which 31 consented to enrollment. We focused on 2 pilot patients sampled at only day 1, and 7 patients sampled serially across 4–11 days that exhibited predominant P. aeruginosa growth in both samples. b Sampling sputum and stool across patients (y-axis) over time (x-axis) from the onset of symptoms. Day 1 sputum sample (teal box) were collected in all patients, and a follow-up sputum (dark blue box) were collected in 7 patients between day 5 and day 12, i.e. 4–11 days after day 1. Stool samples (brown box) with confirmed P. aeruginosa growth were collected in 2 patients. Asterisk: patients with prior P. aeruginosa infection. Treatment with anti-pseudomonal antibiotics are indicated by horizontal lines: piperacillin/tazobactam (weighted black), cefepime (thin black), ceftazidime (dotted black), ciprofloxacin (dotted blue), meropenem (weighted pink). c Samples (sputum or stool) were cultured on cetrimide agar in serial dilutions. A single isolate from day 1 sputum of each patient was used to construct a patient-specific reference genome using long-read sequencing. From each sample, 24 isolates were randomly selected for short-read whole-genome sequencing and phenotyping.