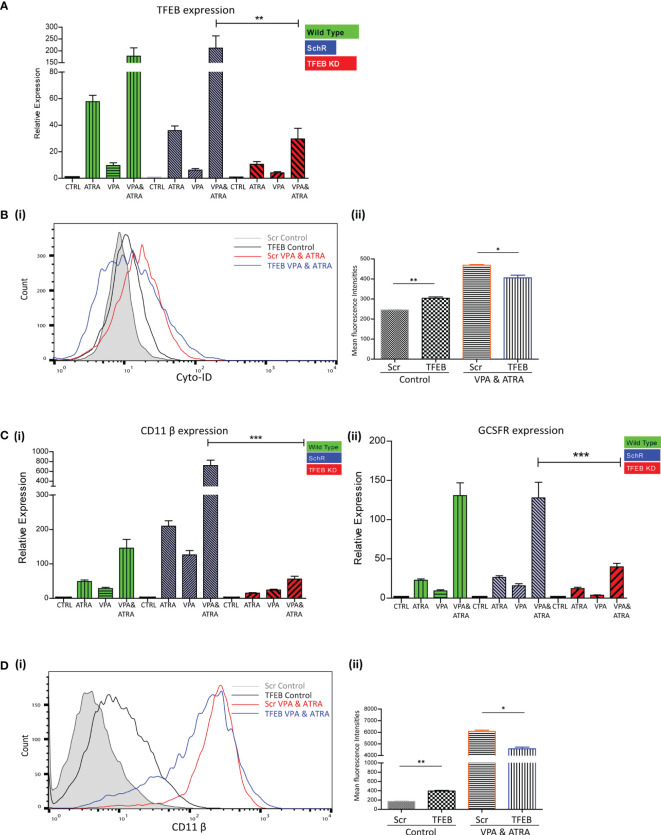
Figure 4.
TFEB knockdown attenuated valproic acid (VPA) & all-trans-retinoic acid (ATRA)-induced autophagy and differentiation. TFEB was knocked down in NB4 cells using lentiviral transduction of target-specific short hairpin (sh)RNA (TFEB). NB4 cells were also transduced with an off-target scrambled shRNA (Scr). Both cell lines were treated with either ATRA (1 μM) or VPA (1mM) alone or a combination of both for 72 hours. (A) Expression levels of TFEB were assessed by RT-qPCR, in wild type (green bars), Scr (blue bars) and TFEB KD (red bars) cell lines. Raw Ct values were normalised to a housekeeping gene and data are shown as n-fold induction compared to untreated controls for each cell line (n = 3) **p < 0.005. (B) (i) Cyto-ID was used to assess autophagosome formation in untreated Scr (grey histogram), untreated TFEB knockdown (black overlay), VPA&ATRA treated Scr (red overlay) and TFEB knockdown (blue overlay) cell lines. (ii) Data from three independent experiments is presented in the graph to the right, as mean fluorescence intensities ± SEM **p < 0.005, *p < 0.05. (C) Expression levels of known differentiation genes CD11β (i) and GCSFR (ii) were assessed by RT-qPCR, in wild type (green bars) Scr (blue bars) and TFEB KD (red bars) cells. Raw Ct values were normalised to a housekeeping gene and data are shown as n-fold induction compared to untreated controls for each cell line (n = 3) ***p < 0.0005. (D) (i) The induction of differentiation was assessed by measuring expression of surface CD11β by flow cytometry in untreated Scr (grey histogram), untreated TFEB KD (black overlay), and VPA&ATRA treated Scr (red overlay) and TFEB KD (blue overlay) cells. A single representative histogram is shown, with mean fluorescence intensity ± SEM presented in the graph to the right (ii) (n = 4) **p < 0.005, *p < 0.05.