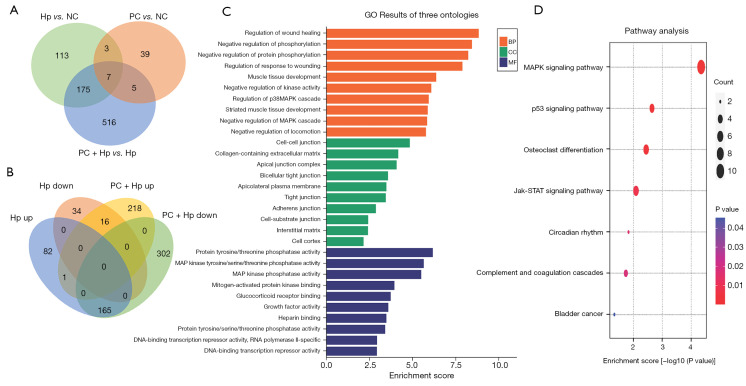
Figure 3.
Transcriptome analysis of Hp-infected AGS cells subjected to phycocyanin intervention. (A) Comparison of differential gene expression profiles between four groups of cell samples: (I) NC vs. Hp; (II) NC vs. PC; and (III) Hp vs. PC + Hp; (B) Venn diagram of genes that were up-regulated during Hp infection versus down-regulated during infection post-phycocyanin intervention, and genes that were downregulated during Hp infection versus up-regulated during infection post-phycocyanin intervention (Hp up represents the number of genes that were up-regulated in Hp infection and Hp down represents the number of genes that were down-regulated in Hp infection. PC + Hp up represents the number of genes that were up-regulated in the PC + Hp group and PC + Hp down represents the number of genes that were down-regulated in the PC + Hp group) (fold change ≥2.0, P<0.05). (C) GO analysis of DEGs. The horizontal axis indicates the enrichment ratio; the larger the enrichment ratio, the stronger the enrichment levels. The vertical axis indicates different pathways. (D) KEGG pathway analysis of DEGs. The highly enriched pathways were screened according to P<0.05 and FDR <0.05. The horizontal axis indicates the enrichment ratio. The vertical axis indicates different pathways. The size of the points indicates the number of enriched genes. NC, AGS cells not infected with Hp nor treated with phycocyanin; Hp, AGS cells infected with Hp only; PC, AGS cells treated with phycocyanin only; PC + Hp, phycocyanin-intervened and Hp-infected cells; GO, Gene Ontology; DEGs, differentially-expressed genes; FDR, false discovery rate.