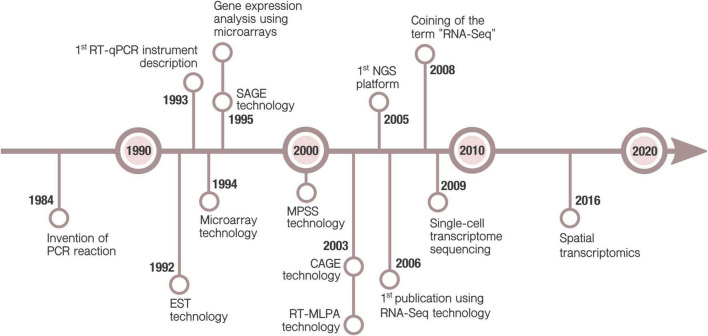
FIGURE 1.
Timeline of main milestones in technologies for gene expression. Polymerase Chain Reaction (PCR) was invented in 1984 by Mullis (1990). A decade later, Real-Time qPCR (RT-qPCR) technology enabled the detection of PCR products in real-time (Higuchi et al., 1993). This allowed, for the first time, quantify mRNA expression of selected genes. Simultaneous analysis of multiple genes was possible due to microarray, Expressed Sequence Tag (EST) (Okubo et al., 1992), Serial Analysis of Gene Expression (SAGE) (Velculescu et al., 1995), Massively Parallel Signature Sequencing (MPSS) (Brenner et al., 2000), Cap Analysis of Gene Expression (CAGE) (Shiraki et al., 2003) and Reverse-transcriptase multiplex ligation-dependent probe amplification (RT-MLPA) (Eldering et al., 2003) technologies. Although the DNA hybridization method was described earlier, the microarray technology is considered to be firstly commercialized by Affymetrix in 1994 (Lenoir and Giannella, 2006). It was promptly applied to measure gene expression (Schena et al., 1995). It continues to be one of the most popular methods for gene expression, allowing the analysis of hundreds or thousands of genes. Development of Next Generation Sequencing (NGS) in the onset of 2000s supposed a revolution for both genomics and transcriptomics (Mardis, 2011). The first publication using NGS RNA-Seq technology was in 2006 (Bainbridge et al., 2006), but it was not until 2008 when the term RNA-seq started to be used (Lister et al., 2008; Mortazavi et al., 2008). RNA-Seq was applied to single-cell technology in 2009 for the first time (Tang et al., 2009). Emerging applications, as spatial transcriptomics (Ståhl et al., 2016), hold the promise of new advances in transcriptomics research.