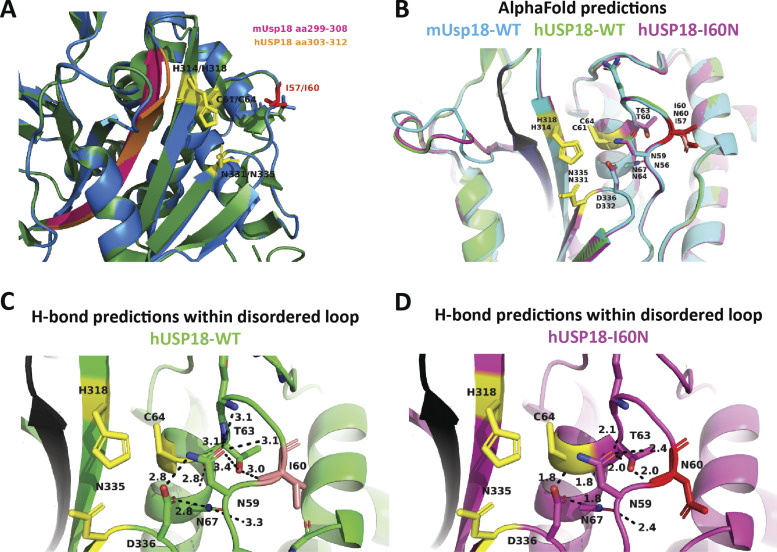
Figure S2.
Prediction modeling of human USP18-WT and I60N variant by SWISS-MODEL and AlphaFold. (A) Overlay of unbound mUsp18 (green) and predicted structure of hUSP18 (blue) modeled by SWISS-MODEL. Residues of the catalytic cleft are shown in yellow, I57/I60 highlighted in red, and residues of β-sheet essential for STAT2 binding highlighted in pink and orange for murine and human USP18, respectively. PDB entry: 5CHT. (B) Alignment of AlphaFold-predicted structures of mUsp18 (cyan), hUSP18 (green), and hUSP18-I60N (magenta) in non–ISG15-bound state. Residue numbering corresponds to human/murine residues in USP18. Residues of the catalytic cleft are shown in yellow. I60/I57 highlighted in salmon/red. Potential STAT2 binding β-sheet is conserved and highlighted in black. (C and D) Polar residue contact prediction within and around the disordered loop. H-bonds formed within the disordered loop of human USP18-WT (C) and USP18-I60N (D) set at a cutoff at 3.5 Å. Figure made using PyMOL.