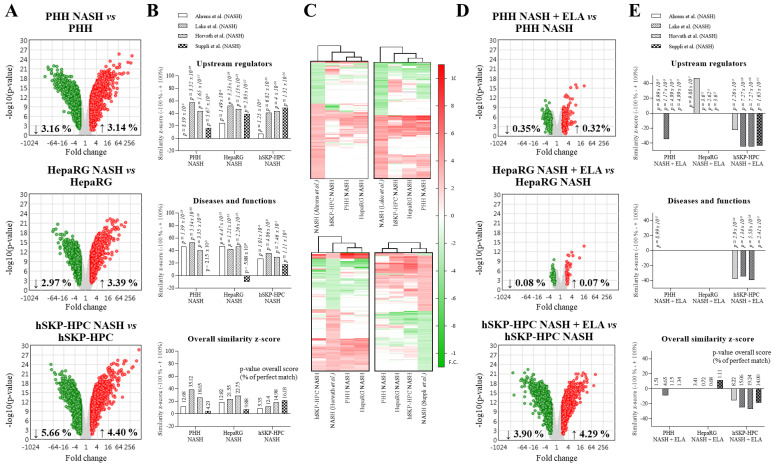
Figure 1.
Transcriptional responses of human hepatic in vitro models exposed to ‘NASH’ triggers correspond with those of human NASH liver biopsies. (A) Microarray volcano plots of differentially modulated probesets in PHH, HepaRG, and hSKP-HPC cultures exposed to ‘NASH’ triggers. (B) Similarity analysis (IPA z-score) of in vitro NASH models and human liver NASH datasets. (C) Hierarchical clustering of upstream regulators in the in vitro NASH models with human liver NASH datasets. (D) Volcano plots of probesets of PHH-, HepaRG- and hSKP-HPC-‘NASH’ cultures exposed to elafibranor. (E) Similarity analysis (IPA z-score) of in vitro NASH models exposed to elafibranor with human liver NASH datasets [Volcano plots exclusion criteria: eBayes, FDR fold change <−2 or >+2 and p < 0.025; IPA analysis: Fisher’s Exact Test, fold change cutoff: <−2 or >+2 and p < 0.025].