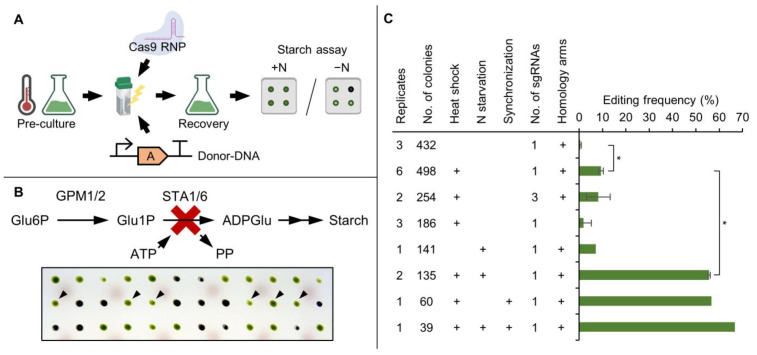
Figure 1.
Optimizing Cas9-RNP-based editing frequency using a phenotypical STA6 knockout screening. (A) Transformation experiments started with a specific pre-culture treatment, followed by electroporation of Cas9-sgRNA ribonucleoproteins (RNPs) in combination with a repair template DNA carrying the aphVII gene. After 24 h of recovery, cells were plated, selected on antibiotic containing TAP agar plates and emerging colonies were transferred onto plates with and without nitrogen (−N/ +N) for starch assay. (B) Pathway of starch synthesis in C. reinhardtii and phenotypical screening via starch assay: Functional knockout of the STA6 gene (red cross) disrupts ADP-glucose formation leading to a starchless phenotype and allowing for visual detection of knockout mutants after exposition to iodine gas (unstained colonies, indicated by black triangles in the second row). (C) Effects of the indicated parameters on editing frequency of the STA6 locus as determined via starch assay. Replicates indicate the number of individual transformations with the total number of colonies screened. Error bars represent the standard deviation of the replicates. Asterisks (*) indicate the significance level of an unpaired, two-sided Student’s t-test assuming non-homogenous variances (*: p < 0.05). Parts of this figure were created with BioRender.com. (A: Antibiotic resistance, GPM: Phosphoglucomutase, Glu6P: Glucose-6-Phosphate, Glu1P: Glucose-1-Phosphate, STA1/6: Adenosine diphosphate glucose pyrophosphorylase, ADPGlu: Adenosine diphosphate glucose, ATP: Adenosine triphosphate, PP: Pyrophosphate).