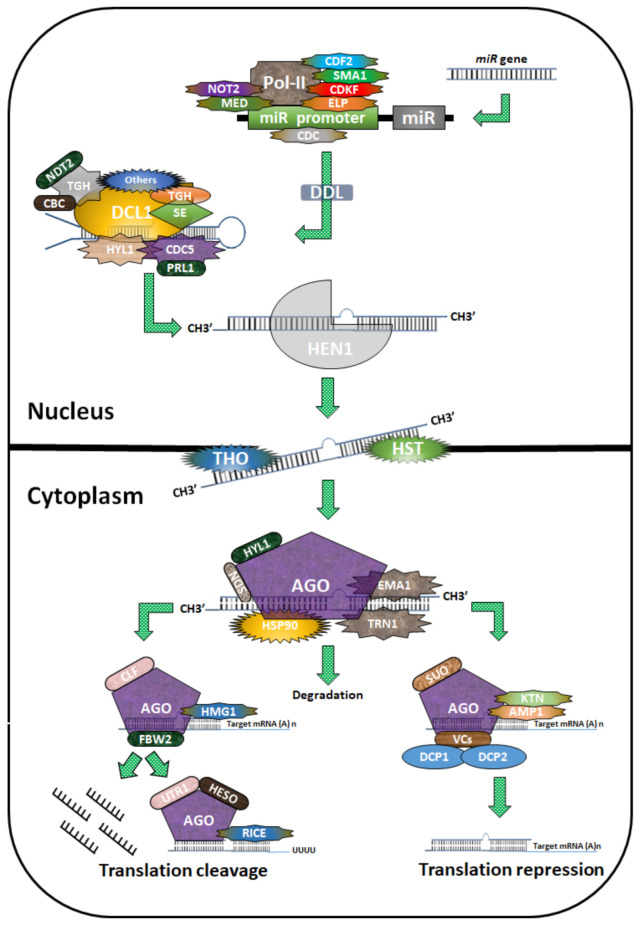
Figure 1.
Biogenesis of miRNAs in plants. The diagram depicts miRNA biogenesis in plants. Pol II first transcribes miR genes into pri-miRNAs, leading to the formation of a hairpin structure. The process is regulated by cycling DOF transcription factors (CDF2), SMALL1 (SMA1), cyclin-dependent protein kinase (CDKF), extension gene (ELP), a protein containing the MYB domain (CDC), Mediator2 (MED2), and NOT2. During nuclear splicing and processing, the cap-binding protein complex (CBC including CBP20 and CBP80HYL1), PRL1 (an evolutionarily conserved WD-40 protein), DDL, TGH, and SERRATE (SE) paly regulatory roles. Dicer-Like 1 (DCL1) progressively processes pri-miRNAs and pre-miRNAs to produce one or more phased miRNA/miRNA* duplexes, which are methylated by HUA enhancer 1 (HEN1) and delivered to the cytoplasm by HST1 (HASTY). The miRNA is chosen and integrated into a specific argonaute1 (AGO1)-containing RISC (RNA-induced silencing complex), which guides translation inhibition or cleavage of the target mRNA transcript.