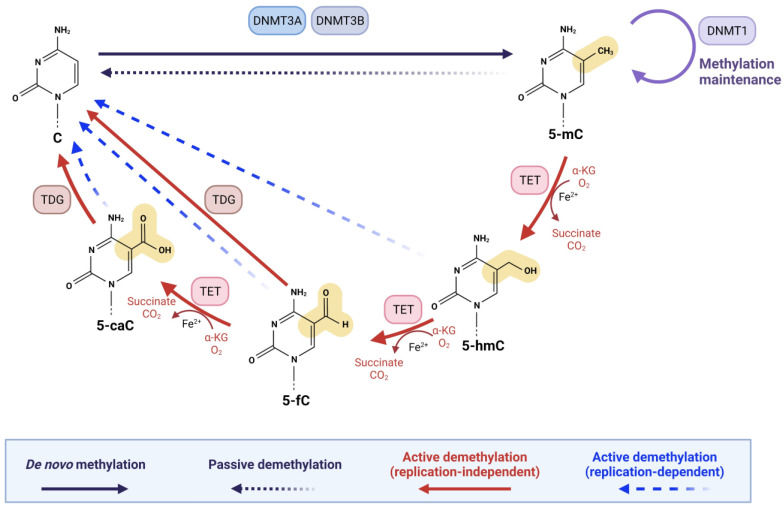
Figure 1.
DNA demethylation in mammals. The two distinct mechanisms of erasure of 5-mC from the mammalian genome, also known as ‘DNA demethylation’, are shown. (1) In active demethylation, TET enzymes (TET1, TET2, and TET3) stepwise oxidize 5-mC to 5-hmC, 5-fC, and 5-caC. 5-fC and 5-caC are then excised by TDG and processed by BER, resulting in reversion of 5-mC to cytosine. Alternatively, replication-dependent dilution of 5-mC oxidation products takes place when 5-hmC, 5-fC, and 5-caC are reversed to unmodified cytosine during DNA replication (see Section 1.2 for detailed description). (2) In passive demethylation, failure of DNMTs function results in replacement of 5-mC by cytosine and dilution of 5-hmC content during DNA replication. 5-caC = 5-carboxylcytosin; 5-fC = 5-formylcytosine; 5-hmC = 5-hydroxymethylcytosine; 5-mC = 5-methylcytosine; α-KG = α-ketoglutarate; BER = base excision repair; C = cytosine; DNMT = DNA methyltransferase; TETs = ten-eleven translocation enzymes (TET1, TET2, and TET3); TDG = thymine DNA glycosylase. Figure was created using the software by BioRender (https://biorender.com/, accessed on 8 February 2022).