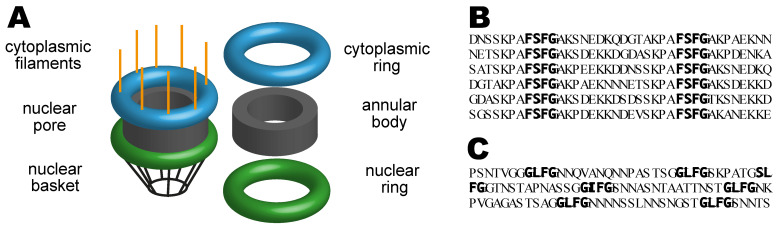
Figure 1.
(A) Highly schematic illustration of nuclear pore structure. Nuclear pores span the double membrane of the envelope and are based on a central annular core (sometimes referred to as a central ring [10] and which is constructed from two similar halves) that is sandwiched between inner (nucleoplasmic) and outer (cytoplasmic) rings. Appendages also extend into the cytoplasm and the nucleus (for clarity these have been omitted from the exploded view). Complementary X-ray crystallography and cryo-EM studies have established the arrangement of the nuclear pore proteins (nucleoporins or “nups”) in S. cerevisiae and other species [1,2,3,4,5,6,7,8,9,10]. Nuclear pores serve as a conduit between the nuclear and cytoplasmic compartments, with macromolecules such as proteins and RNAs moving through the central channel. Many nucleoporins (“FG-nucleoporins”) contain long regions containing characteristic sequence repeats based on cores rich in Phe (F) and Gly (G) that lack secondary structure (or are “natively unfolded”) and which protrude into the central channel where they form a dense mesh that impairs the movement of macromolecules. The fibrous nuclear basket and cytoplasmic filaments have been omitted for clarity in the exploded image. (B) Portion of the S. cerevisiae FG-nucleoporin Nup1 showing FG rich cores (bold), here containing FSFG (bold), interspersed with linkers of variable sequences that generally lack hydrophobic residues. (C) Portion of the sequence of S. cerevisiae Nup116 that contains cores based on a GLFG motif (bold). Here the lengths of the regions between the FG cores are more variable than in Nsp1.