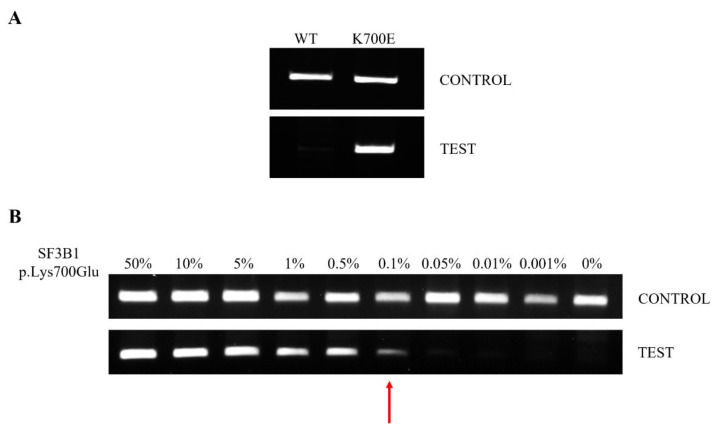
Figure 2.
Electrophoretic runs of PCR reactions: each amplicon was loaded on 2% agarose-TBE 1× gel with 5 μg/mL EtBr and run at 150 V for 30 min. PNA-PCR clamping for SF3B1 was carried out in the absence (CONTROL) and presence (TEST) of the PNA probe. (A) Example of PNA-PCR clamping in DNA from WT and p.Lys700Glu-mutated patients. CONTROL PCR represents an internal control and DNA must always be amplified. TEST PCR returns the result about the mutational status of SF3B1; if the patient is SF3B1 WT, the gDNA is not amplified, while if the patient is SF3B1 p.Lys700Glu, the gDNA is amplified. (B) PNA-PCR clamping LoD was assessed by mixing pGEMT-SF3B1 p.Lys700Glu and pGEMT-SF3B1 WT plasmids at different ratios in the same PCR reaction. Dilutions were as follows: 50, 10, 5, 1, 0.5, 0.1, 0.05, 0.05, 0,01, 0.001 and 0% pGEMT-SF3B1 p.Lys700Glu, all brought to 100% with the respective amount of pGEMT-SF3B1 WT template. The percentage of the mutated template is indicated above each amplicon. The red arrow indicates the LoD of the PNA-PCR clamping method.