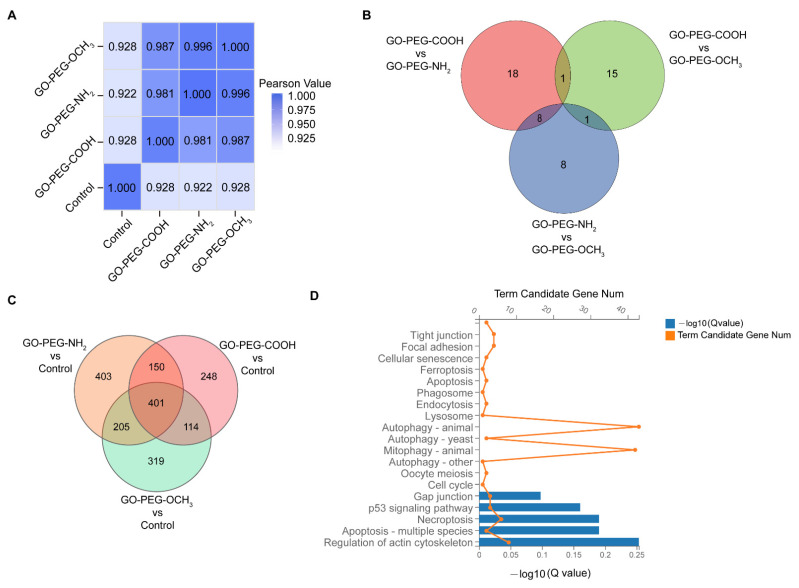
Figure 3.
Gene expression profile analysis of hCorECs exposed to PEG-GO nanosheets. (A) Pearson correlation analysis of all gene expression in hCorECs of four groups. (GO-PEG-COOH vs. GO-PEG-NH2, 0.981; GO-PEG-OCH3 vs. GO-PEG-NH2, 0.996; GO-PEG-OCH3 vs. GO-PEG-COOH, 0.987; GO-PEG-COOH vs. Control, 0.928; GO-PEG-NH2 vs. Control, 0.922; GO-PEG-OCH3 vs. Control, 0.928). (B) Venn diagram showed the differentially expressed genes (DEGs) from paired comparisons among three PEG-GO groups. (C) Venn diagram showed the DEGs from paired comparisons between PEG-GO groups and control group. Genes with |log2 FC (fold change)| > 1 and adjusted p value ≤ 0.05 were considered to be DEGs. (D) The pathway analysis of 401 DEGs in terms of cellular processes based on Kyoto Encyclopedia of Genes and Genomes databases.