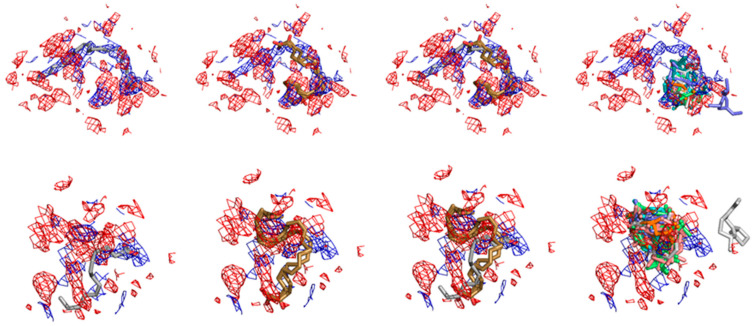
Figure 7.
Different binding modes for the ligands at the COX-1 crystallographic and docked structures and their fits to crystal electronic density. At the Top the electronic densities and different molecular model superpositions for the human COX-1 complexed with arachidonic acid structure at the PDB-ID:1DIY is shown. At the bottom, the superpositions with the electronic density of the same complex as solved at the PDB-ID:1U67 are depicted. In both cases, the red and blue meshes depict the respective FoFc electronic density (i.e., electronic densities for which the authors model have found atomic fitting) and 2FoFc maps (i.e., electronic densities for which the author’s molecular model has not found atomic superposition). In both cases also the respective electronic density maps are considered with a σ factor of 1.0. From left to right it can be seen the respective superposition at both densities for the models deposited at the protein data bank; for our Arachidonic acid docked structures (positive control); for both (crystallographic and docked); for the set of the three first docked poses of the top hits for this enzyme between the VOCs profiled from the M. urundeuva seeds.