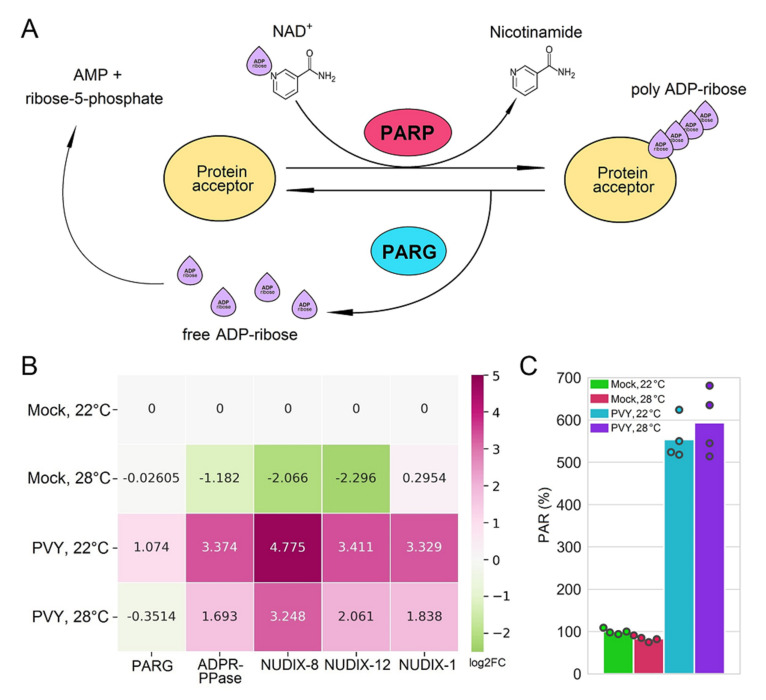
Figure 5.
Poly (ADP-ribose) metabolism and susceptibility of Chicago plants to PVY. (A) Schematic representation PARylation process in healthy cells. Poly(ADP-ribose) polymerase (PARP), poly(ADP-ribose) glycohydrolase (PARG), and nucleotide diphosphate linked to some moiety-X (NUDIX) enzymes. PARP enzymes bind NAD+ (nicotinamide adenine dinucleotide), cleave off the nicotinamide residue, and attach the remaining ADP-ribose moiety to acceptor proteins (protein X, which can include PARP itself). PARG then cleaves the ribose–ribose backbone bond of poly(ADP-ribose), releasing free ADP-ribose. ADP-ribose-specific NUDIX enzymes then cleave free ADP-ribose into AMP (adenosine monophosphate) and ribose-5-phosphate. (B) Heatmap showing the changes in abundance (log2FC) of mRNAs related to the poly ADP-ribosylation process: PARG (Soltu.DM.12G003820.1), ADPR-PPase (ADP-ribose pyrophosphatases; Soltu.DM.08G000850.1) and NUDIX enzymes (Soltu.DM.03G005230.1-NUDIX-1; Soltu.DM.08G000940.1—NUDIX-12; Soltu.DM.08G000920.1-NUDIX-8). (C) Accumulation of PARylated proteins measured by ELISA using rabbit anti-PAR polyclonal antibody, in PVY- systemically infected or mock-inoculated plants at 22 °C or 28 °C. Analysis of variance and Tukey’s HSD post hoc tests were performed for data.