Fig. 1.
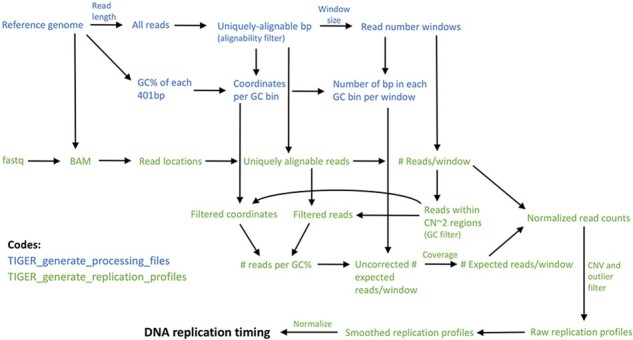
TIGER pipeline overview. TIGER consists of two scripts, the first run once for a given reference genome and window size, and the second applied per sequenced sample. TIGER extracts read locations from a sequencing library, filters reads for alignability, counts reads in genomic windows of uniform alignability, calculates a GC bias factor and uses it to correct the window read counts, filters for CNVs and outliers, and smoothes and normalizes the data to derive the final DNA replication timing profiles
