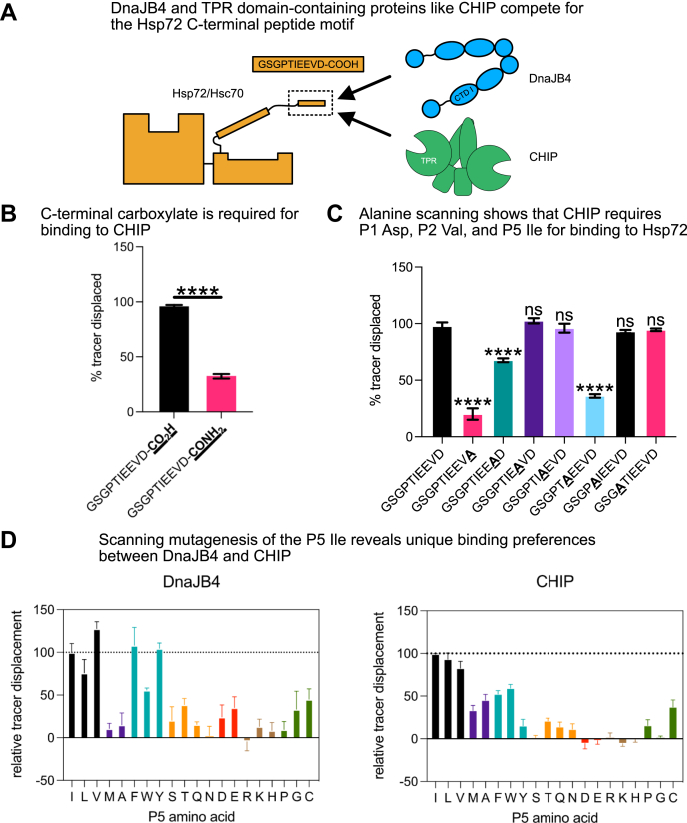
Figure 5.
DnaJB4 accommodates an expanded number of amino acids at P5, compared with CHIP.A, cartoon depicting competition for the Hsp70 C-terminal EEVD motif by DnaJB4 and CHIP. B, competition fluorescence polarization (FP) experiment comparing displacement of 20 nM Hsp72 tracer from 1.58 μM CHIP by 100 μM WT or C-terminally amidated Hsp72 peptides. Graph shows mean tracer displacement relative to dimethyl sulfoxide (DMSO) control ±SD (n = 4). Statistics were performed using unpaired Student’s t test (∗∗∗∗p < 0.0001 compared with WT control). C, competition FP experiment comparing alanine scanning substitutions across the Hsp72 C-terminal sequence in binding to CHIP. Graph shows mean tracer displacement relative to DMSO control ±SD (n = 4). Statistics were performed using unpaired Student’s t test (∗∗∗∗p < 0.0001, ns, not significant compared with WT control). D, competition FP experiment comparing all possible mutations at the P5 position of the Hsp72 EEVD motif in binding to DnaJB4 or CHIP. DnaJB4, 5 μM, or CHIP, 1.58 μM, was incubated with 20 nM WT Hsp72 tracer and 100 μM unlabeled competitor peptide. Tracer displacement was calculated relative to DMSO control and normalized to WT as 100% displacement. Graph shows mean relative tracer displacement ±SD (n = 4).