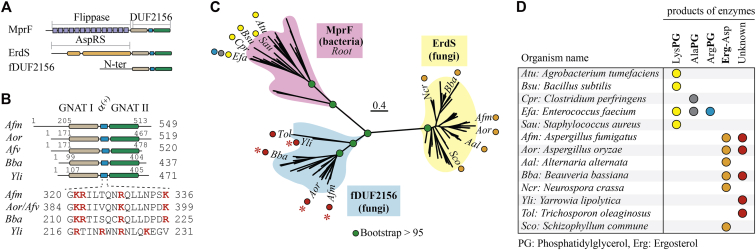
Figure 1.
In silico characterization of fungal fDUF2156 proteins.A, architecture of bacterial aaGLSs (MprFs), with the N-terminal aaGLs flippase (14 transmembrane helices) and the DUF2156 domain. Fungal ErdS (AspRS-DUF2156) are represented as well as fungal fDUF2156 proteins. B, fDUF proteins from Afm, Aor, Afv, Bba, and Yli. The two Gcn5-N-acetyltransferase (GNAT) domains separated by the α(+) helix, which constitute the DUF2156 fold, are detailed (domain coordinates indicated, length of proteins on the right). K (Lys) and R (Arg) residues are shown in red. C, maximum likelihood phylogenetic reconstruction of bacterial (MprF) and fungal (ErdS, fDUF) DUF2156 domains. Three clades are visible that match bacterial MprF- (purple background), ErdS- (yellow background), and fDUF2156- (blue background) specific groups. Positions of some proteins are indicated with the abbreviated name of the organism in which they are found together with (D) the aminoacylated products that they synthesize (if known). Red asterisks indicate the proteins studied in this work.