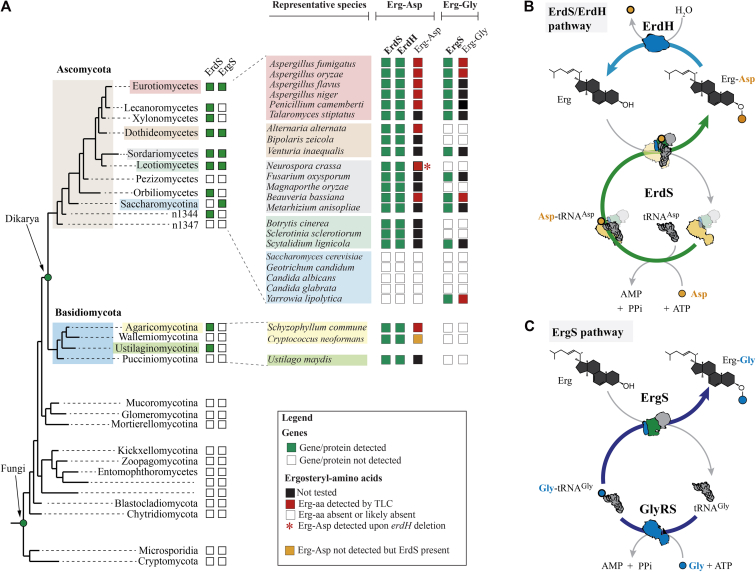
Figure 7.
Distribution of ergosteryl-amino acid synthases in fungi, and overview of Erg-Gly and Erg-Asp synthesis mechanisms and factors. ErgS (Erg-Gly synthase) and ErdS (Erg-Asp synthase) sequences were searched across eukaryotes and, most specifically, in the fungal kingdom, and the presence (green squares) or absence (white squares) of the corresponding proteins was mapped onto a simplified version of the phylogenetic tree of fungi (A), as provided by the JGI Genome Portal (see Experimental procedures), in main fungal lineages. In Eurotiomycetes (red), Dothideomycetes (light brown), Sordariomycetes (gray), Leotiomycetes (light green), and Saccharomycotina (blue), the presence/absence of ErgS and ErdS was indicated in several representative species (as well as in three basidiomycetes in Agaricomycotina and Ustilaginomycotina). Erg-aa synthases are detected only in Dikarya and ErgS only in Ascomycota. B, Erg-Asp biosynthesis and degradation pathway. ErdS uses free L-Asp, ATP, and tRNAAsp and produces Asp-tRNAAsp in its AspRS domain (yellow), which is transferred to the appended DUF2156/ATT domain where it serves as the Asp donor for Erg-Asp synthesis. ErdH is an Erg-Asp specific hydrolase (whose distribution is indicated in (A)). C, Erg-Gly synthesis pathway. GlyRS uses Gly, ATP, and tRNAGly to produce Gly-tRNAGly, which is then released, captured by ErgS, and used as the Gly donor for Erg-Gly synthesis.