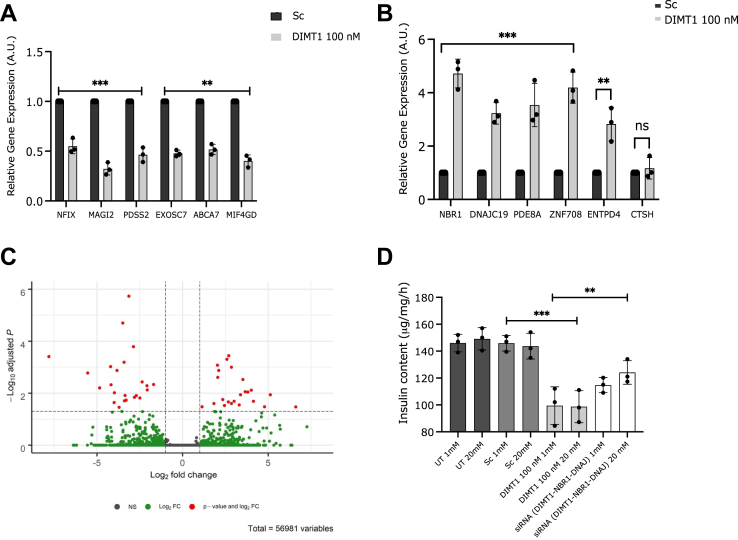
Figure 6.
Validation of RNA sequencing by quantitative real-time PCR. Differentially expressed genes (DEGs) identified by RNA sequencing were evaluated; a total of six DEGs were selected based on their (i) fold change, (ii) relevance to cellular pathways, and (iii) expression levels (A and B; data are mean ± SD; n = 3). C, volcano plot representing significantly downregulated genes (log2fold change < (−1) and adjusted p-value < 0.05), which are indicated in red on the left side of the plot, while significantly upregulated genes (log2fold change > (1) and adjusted p-value < 0.05) are indicated in red on the right side of the plot. Vertical dashed lines correspond to the log2 fold change threshold of |1| and horizontal dashed line corresponds to the adjusted p-value threshold of 0.05 represented as log10 adjusted p-value. D, changes in insulin content in NBR1-and DNAJC19-silenced cells. The graphical representation of the qRT-PCR analysis and the data are expressed as mean ± SD (n = 3). Statistical analysis was done using paired Student’s t test. ∗∗p < 0.01, ∗∗∗p < 0.001, NS, non-significant.