Graphical abstract
Keywords: MPEPE, Deep learning, Protein expression, Mutation
Highlights
-
•
Gene sequence is the major factor contributing expression.
-
•
MPEPE is a mutation predictor for enhancing protein expression.
-
•
Identified mutations in enzymes increased its expression levels in E. coli.
Abstract
The expression of proteins in Escherichia coli is often essential for their characterization, modification, and subsequent application. Gene sequence is the major factor contributing expression. In this study, we used the expression data from 6438 heterologous proteins under the same expression condition in E. coli to construct a deep learning classifier for screening high- and low-expression proteins. In conjunction with conserved residue analysis to minimize functional disruption, a mutation predictor for enhanced protein expression (MPEPE) was proposed to identify mutations conducive to protein expression. MPEPE identified mutation sites in laccase 13B22 and the glucose dehydrogenase FAD-AtGDH, that significantly increased both expression levels and activity of these proteins. Additionally, a significant correlation of 0.46 between the predicted high level expression propensity with the constructed models and the protein abundance of endogenous genes in E. coli was also been detected. Therefore, the study provides foundational insights into the relationship between specific amino acid usage, codon usage, and protein expression, and is essential for research and industrial applications.
1. Introduction
High-level production of soluble recombinant proteins at low cost is crucial for their application in many fields. However, there are barriers to heterologous expression in many hosts, including in the unicellular bacterial Escherichia coli expression system. In order to minimize the effect of heterologous recombinant proteins on cell growth, a smart expression host could degrade the heterologous proteins by the proteolysis system or form an inclusion body. It is estimated that <50% of bacterial and <15% of non-bacterial proteins could be expressed in soluble form in E. coli [1].
Studies of expression system modification have examined, for example, the optimization of expression conditions [2], [3], [4], [5], expression of co-soluble tags fused to target proteins [6], as well as co-expression of the molecular chaperone [7]. While these methods work for a small number of proteins, the level of improvement for most proteins is still limited and some strategies to increase expression reduce the catalytic activity.
Additional studies show that there are many different strategies to increase the soluble and functional expression of foreign proteins. According to Deng et al., alanine- or leucine-scanning mutagenesis increased soluble expression, and leucine could increase the protein helices and their stability against degradation by proteinase K [8]. In recent years, the well-established directed evolution approach has been employed to optimize the coding sequence based on soluble expression phage-assisted continuous evolution (SE-PACE). This method has been used to evolve some antibody fragments and maltose-binding protein (MBP) to increase their expression [9]. Michal Jamroz et al. conducted a promising rational design study in which they proposed the AGGRESCAN 3D structural aggregation predictor. This method was used to modify the green fluorescent protein and the human single-domain VH antibody, effectively reducing the aggregation propensity of the protein and increasing its expression [10]. In summary, those results indicated that the sequence is also a vital factor to regulate its expression.
One promising approach for uncovering the key factors affecting high-level and soluble expression is deep learning, a subfield of machine learning that uses multi-layered Deep Neural Networks (DNNs) to extract novel features from input data [11]. The first predictive tool for protein solubility based on amino acid sequence was proposed by Wilkinson and Harrison in 1991 [12]. Based on 81 soluble or insoluble protein sequences, the authors found that the charge average and turn-forming residue fraction correlated with inclusion body formation. In recent years, deep learning algorithms for enhancing the expression of exogenous proteins in E. coli have been developed. Approaches based on logistic regression [13] and deep learning architecture (EPSOL) [14] were proposed to learn the features related to soluble expression from the expressed sequence fragments and construct the classifier to distinguish the soluble expressed proteins, but they lacked the experimental validation. A model based on the bidirectional long-short-term memory conditional random field was constructed to optimize the codon and enhance protein expression [15]. A feed-forward artificial neural networks were also constructed based on the ribosome profiling [16]. All of the results indicated that the gene sequence could affect the protein expression. A complicated model should be trained based on the real gene sequence and design the gene to improve its expression.
In this study, we propose a predictive model, mutation predictor for enhanced protein expression (MPEPE). The 6438 proteins that were experimentally validated the expression yields in E. coli [17] were selected to train and validate the prediction model based on multi-layered deep neural networks (DNNs). The evolutionary method was incorporated into the model to virtually screen the mutant sites that might make positive contributions to protein expression but not disrupt its function. When the strategy was applied on two enzyme proteins, the laccase 13B22 and the flavin adenine dinucleotide-dependent glucose dehydrogenase (FAD-AtGDH), the expression and activity of these two enzymes in E. coli were significantly increased. This study will help researchers to understand the relationship between amino acids and soluble protein expression and is important for the industrial application of enzymatic proteins.
2. Materials and methods
2.1. Collection of the protein expression dataset
We collected the protein expression dataset from a published study [17]. All of the 6438 proteins in the dataset have been classified to six classes (Class1, Class2, Class3, Class4, Class5, and Class6) based on the protein expression level under the identical conditions and from the same promoter in E. coli in the reference [17]. The protein level was scored on the integer scale from class1 (lowest) to class6 (highest) based on the visual inspection of whole cell lysates in Coomassie-blue-stained SDS-PAGE gels [17], [18]. The proteins in Class1 have the lowest protein levels while the proteins in Class6 have the highest protein levels. For the aim to construct the relative balanced training dataset in terms of the size, the low expression dataset consisted of 2308 proteins in Class1, Class2, and Class3, while the 1973 proteins in Class6 comprised the high expression dataset. Therefore, the ratio of low expression proteins to high expression proteins was ∼1:1. The independent validation dataset was composed of the proteins in Class4 and Class5 and its size was 2067. Table 1 shows detailed information on the size of each dataset. The protein abundance data were downloaded from the paxdb database and the weighted average of WHOLE_ORGANISM between the different protein abundance values was taken (http://pax-db.org/) [19].
Table 1.
Datasets classification and its size.
| Dataset | Evaluation Scores | Class | Sequence Number | Constructed Datasets |
|---|---|---|---|---|
| lixiProtein Expression Dataset | 1 | Negative data | 1754 | low expression dataset |
| 2 | Negative data | 131 | ||
| 3 | Negative data | 423 | ||
| 4 | – | 896 | validation dataset | |
| 5 | – | 1171 | ||
| 6 | Positive data | 1973 | high expression dataset |
2.2. Data processing of protein expression datasets and coding schemes
In order to use synonymous codon number, the specific amino acid, and specific nucleotide combination to construct three Deep Neural Network (DNN) models, the nucleotide (codon) sequences in the protein expression dataset were translated into amino acid sequences and codon number sequences, the detailed coding information was shown in Table S1.
We used the one-hot encoding method to code different sequence-style datasets under different coding schemes. The synonymous codon number sequences were transformed by a 6 × 6 matrix; for instance, codon number 1 was encoded by the vector (1,0,0,0,0,0), codon number 6 was encoded by the vector (0,0,0,0,0,1):
The specific amino acid sequences were transformed by a 20 × 20 matrix; for example A (Ala, Alanine) was encoded by vector and Y (Tyr, Tyrosine) was encoded by vector :
Specific nucleotide combination sequences were transformed by a 61 × 61 matrix; for example, the index of “GCT” in Table S1 was 1 and it was encoded by the vector , and the index of “TAT” in Table S1 was 61 and it was encoded by the vector :
2.3. DNN architecture in three models
We used convolution layers, pooling layers, and Long-Short Term Memory (LSTM) layers together to make up the DNN architecture. DNN models were trained with the three constructed datasets based on a 10-fold cross-validation strategy. And the specific process of the 10-fold cross-validation strategy here is as follows. The negative and positive samples are randomly partitioned into k(k = 10) equal size subsamples. Of the k subsamples, a single subsample is retained as the validation data for testing the model, and the remaining k-1 subsamples are used as training data. The 10-cross-validation process is then repeated k(k = 10) times (the folds), with each of the k subsamples used exactly once as the validation data. Based on this method, all samples are used for both training and validation, and each sample is used for validation exactly once. The detailed architectures and optimized hyper-parameters showed in Fig. S1 and three DNN models were trained on the TensorFlow platform [20] based on Keras 2.1.5 in the Python 2.7.15 programming environment.
Three models were successfully constructed. The architecture of each one started with an embedding layer, followed by two convolutional, maximum pooling layers, an LSTM layer, and a final prediction layer. In addition to the prediction layer, each layer was followed again by a Batch Normalization layer and a final dropout layer. In this model, the rectified linear unit (ReLU) activation function was used (except for the final prediction layer) as follows:
where x denotes the feature map from the convolution operation (the weighted sum of a neuron). The softmax activation function was used in the final prediction layer.
To avoid over-training and the resultant over-fitting of the prediction model, we used an early stopping technique to detect the prediction accuracy achieved the high score on the test dataset in the training process. We optimized the various hyper-parameters in the DNN architecture, including the number of layers, number of kernels, kernel size, fully connected layer size, dropout rate, learning rate, batch size, activation functions, number of nodes, and optimizers using the optimization package HYPERAS (https://github.com/maxpumperla/hyperas). The final parameters used for the prediction models are shown in Table S2. We also verified the performance of the proposed method by a 10-fold cross-validation method. The code for the above DNN architecture with Python v2.7.15 environments is available from GitHub (https://github.com/BRITian/MPEPE).
2.4. Evaluation of prediction performance
A 10-fold cross-validation strategy was used to train and evaluate the performance of the prediction models. The performance of the prediction models was evaluated with metrics including accuracy, recall, precision, F1-score, and the area under the receiver operating characteristic curve (AUROC) score, which were each calculated based on 10-fold cross-validation. The area under ROC (AUROC) and the area under PRC (AUPRC) of the prediction models were greater than 0.5, which indicated that the performance of the constructed models was better than that of a random classifier. The metrics were calculated using the Keras package as follows:
where TP, TN, FP, and FN indicate true positive, true negative, false positive, and false negative, respectively.
2.5. The PSAP and entropy analysis
Using BLASTP, we identified and downloaded sequences of homologous proteins of laccase 13B22 and the glucose dehydrogenase FAD-AtGDH (identity >40% and coverage >50%), from NCBI (https://www.ncbi.nlm.nih.gov/). The homologous protein sequences were aligned with their respective wild type and spliced to the same length. Then the position-specific amino-acid probability (PSAP) matrices for both enzymes were calculated to determine the conservation of amino acids at each locus. The entropy of each residue was then determined using the PSAP calculation tool [4], [21], [22]. In this study, we used the cutoffs of PSAP value of mutations greater than 0.1 and the entropy value of the residue greater than 0.65 to select the non-conserved residues and natural selected mutations.
2.6. Simulation of protein three-dimensional structure
The AlphaFold 2.0 tool was used to simulate the three-dimensional structure of laccase 13B22 and FAD-AtGDH from its amino acid sequence with the publicly available code and default parameters [23]. All five structures were simulated and the structure with the least structural energy was selected for the following analysis.
2.7. Construction of mutants
We generated single-point mutations in the residues that differed from the sequence of the wild type in the mutants using the two-step PCR mutagenesis strategy [24]. Mutant primers were designed using Oligo software ver.7.0 (Table S3) and were then used to generate the single-point mutants. Using wild-type plasmid DNA template, we amplified the mutant site by T7-F primer and each downstream primer containing a mutation sequence or T7-R and each upstream primer containing a mutation sequence under the action of Phanta Max Super-Fidelity DNA Polymerase (Vazyme, Nanjing, China). PCR was performed as follows: 95 °C for 5 min for the preheat, 95 °C for 30 s, 58 °C for 30 s, 72 °C for 1 min, 32 cycles, and 72 °C for 10 min. We used an AxyPrep DNA gel extraction kit (Axygene, California, USA) to recover the target gene fragment, and then it was used as the primer and the wild-type plasmid as the template for the second round of PCR amplification. The mutant plasmid was obtained by PCR amplification. The second round PCR was performed as follows: 95 °C, 5 min for the preheat, 95 °C for 30 s, 72 °C for 5 min, 32 cycles, and 72 °C for 10 min. The wild-type plasmid was eliminated by DpnI (NEB, Ipswich, United Kingdom) and the mutant plasmids were recovered using a purification and recovery kit (TransGen, Beijing, China) and then transformed into E. coli Top10 competent cells (TransGen, Beijing, China) using standard procedures [25]. Through bacterial liquid PCR, the single clone with the correct size of the target gene was preliminarily identified and sequenced by TSINGKE biological technology (Beijing, China), and then the correct mutant plasmid was transformed into competent E. coli BL21 (DE3) cells (TransGen, Beijing, China). Multi-site mutations were obtained by adding two mutation sites in each round according to the above method.
2.8. Detection of the expression of laccase 13B22
E. coli BL21 (DE3) cells harboring the recombinant plasmid were cultured in a 50 mL LB (Lysogeny Broth) medium supplemented with kanamycin (50 μg/mL) at 30 °C with shaking at 120 rpm to an OD600 of 0.7∼0.75. Then, 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and 0.4 mM CuCl2 were added to the culture medium, and the temperature was reduced to 25 °C. Incubation was continued for a further 4 h, during which microaerobic conditions were achieved by switching off the shaking function [26], [27]. Cells were harvested after a further 20 h of growth by centrifugation at 8000g for 10 min [28]. The pellets were resuspended in 5 mL buffer containing 20 mM Tris-HCl (pH 8.0). The cells were disrupted by sonication on ice, and debris was removed by centrifugation at 4 °C and 8000g for 30 min. After transferring the crushed supernatant to a new pre-cooled 10 mL EP tube and resuspending the crushed pellet in 5 mL of 20 mM Tris-Cl buffer (pH 8.0), the laccase activity was determined. 50 µL protein were taken concurrently to prepare SDS-PAGE samples.
SDS-PAGE was performed using standard procedures and gels were stained with Coomassie brilliant blue R250. For Western blotting analysis, SDS-PAGE gels were transferred to polyvinylidene fluoride (PVDF) membranes (Amersham, Piscataway, NJ, USA), and these were blocked with 5% non-fat milk (Applygen, Beijing, China) and incubated with mouse anti-His monoclonal antibody (TransGen, Beijing, China), followed by horseradish peroxidase HRP-conjugated goat anti-mouse IgG (TransGen, Beijing, China). Proteins were visualized using a BeyoECL Plus Chemiluminescence Detection Kit (Beyotime, Shanghai, China) and Chemiluminescence Touch Imaging System (e-BLOT, Shanghai, China).
2.9. The detection of the activity of the laccase 13B22
Laccase activity was assayed at 37 °C using 2,2-azino-di-(3-ethylbenzthiazoline-sulfonate) ABTS (Sigma-Aldrich, St. Louis, USA) as substrate. An assay mixture containing 200 µL 5 mM ABTS and 750 µL 50 mM citrate/phosphate was preheated at 37 °C for 2 min, and then 50 µL crushed supernatant was added to react accurately for 3 min. This mixture was then terminated in an ice-water bath for 1 min. The increase in absorbance due to the oxidation of ABTS at 420 nm was measured (ε420 = 36,000 M−1 cm−1). One unit was defined as the amount of enzyme that oxidized 1 μmol of substrate per minute [29].
2.10. Detection of the expression of glucose dehydrogenase activity of FAD-AtGDH
E. coli BL21 (DE3) cells harboring the recombinant plasmid were cultured in a 50 mL LB medium supplemented with kanamycin (50 μg/mL) at 37 °C with shaking at 200 rpm to an OD600 of 0.7∼0.75. Then 0.3 mM isopropyl-β-d-thiogalactopyranoside was added to the culture medium, which was grown for 18 to 20 h in LB medium at 16 °C and 200 rpm to obtain FAD-AtGDH as soluble and active proteins. Cells were harvested by centrifugation at 8000g for 10 min. The pellets were resuspended in 5 mL buffer containing 20 mM Tris-HCl (pH 8.0). The cells were disrupted by sonication on ice, and debris was removed by centrifugation at 4 °C and 8000g for 30 min. The crushed supernatant was then transferred to a new pre-cooled 10 mL EP tube, and the crushed pellets resuspended in 5 mL of 20 mM Tris-Cl buffer (pH 8.0), and the glucose dehydrogenase activity was determined. At the same time, 50 µL protein were taken to prepare SDS-PAGE samples.
2.11. Detection of the activity of glucose dehydrogenase activity of FAD-AtGDH
FAD-AtGDH activity was assayed spectrophotometrically using 2,6-dichloroindophenol (DCIP, ɛ600 = 16.3/mM/cm) (Solarbio, Beijing, China) and phenazine methosulfate (PMS) (Solarbio, Beijing, China) as electron acceptors. The reaction was followed for per min measured and the reaction continued for 5 min at 600 nm using the SpectraMax M2 microplate reader (Molecular Devices, Silicon Valley, USA). The DCIP-based assay contained 50 mM Potassium phosphate buffer (pH 6.5), 0.06 mM DCIP, 0.6 mM PMS and 100 mM D-glucose. One unit of FAD-AtGDH activity was defined as the amount of enzyme required for the reduction of 1 μmol glucose or electron acceptor per min under the assay condition [30].
3. Results
3.1. Overview of the MPEPE prediction strategy
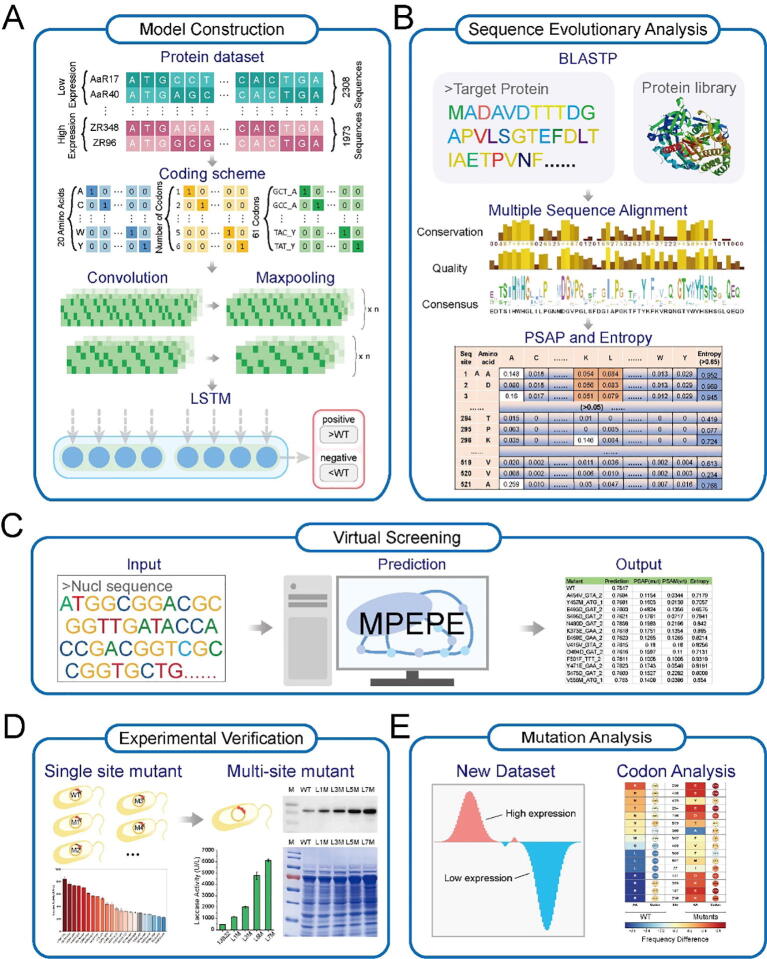
In order to predict mutations that could enhance the heterologous expression of proteins in the soluble fraction in E. coli, we proposed the MPEPE strategy for rational optimization of gene sequence related to translation rate and protein surface charge. To this end, we constructed DNNs, the predicted accuracy of which was higher than the other machine learning methods (Fig. S2, Table S4), to select mutations with a high potential to increase protein expression levels based on published, experimentally-determined expression data generated in E. coli under the identical expression condition and expression system including the promoter (Fig. 1A; and more details in the next sub-section) [17]. In addition, evolutionary analysis was employed to select non-conserved residues for mutagenesis that appeared unlikely to disrupt protein function (Fig. 1B). Based on the above two design components (Fig. 1A and B), we screened single-point mutations that might positively contribute to expression levels, but not disrupt the catalytic function of the protein of interest (Fig. 1C). We experimentally verified the contribution of each screened mutation by SDS-PAGE and enzymatic activity analysis, then generated mutants with stacked mutations to determine the combined effects of these confirmed positive mutations (Fig. 1D). Through this discovery pipeline we obtained several candidate mutations for exploration of the determining factors related to increased protein expression in the soluble fraction in E. coli (Fig. 1E) and to subsequently design a mutant with significantly enhanced expression in E. coli.
Fig. 1.
The workflow of MPEPE based on deep learning and evolutionary analysis. A. The protein datasets were used as inputs for constructing and training the prediction model. B. Mutation sites were screened using evolutionary analysis of target protein sequence without disrupting function. C. The target nucleotide sequences were used as inputs in the MPEPE to virtually screen mutants. D. Experimental validation on the effect of virtual screened mutants on their expression level in E. coli. E. A new data set was constructed based on the experimental results for optimizing the MPEPE model.
3.2. Protein expression data-based selection of charged amino acids and rapidly translated codons in E. coli
To investigate relationships between protein expression level and amino acid charge properties or synonymous codon number for specific amino acids, we identified preferentially selected amino acids/codons in 6438 published proteins that were experimentally classified into six categories based on their heterologous protein expression under identical conditions and promoter in E. coli, collected from diverse phylogenetic sources and provided broad sampling of codon and amino acid space due to variations in codon-usage frequency in the source organisms [17]. The expression indexes for the six classes ranged from 1 to 6 (from lowest to highest), and consisted of n = 1754 proteins in group 1, n = 131 in group 2, n = 423 in group 3, n = 896 in group 4, n = 1171 in group 5, and n = 1973 in group 6. Due to substantial bias in the number of proteins allocated among the six classes, groups 1, 2, and 3 were combined to represent low expression proteins, while group 6 represented highly expressed proteins. The resulting dataset was relatively balanced, with a low:high ratio of 1:0.85. Proteins in groups 4 and 5 were selected as independent categories to validate the performance of the prediction model.
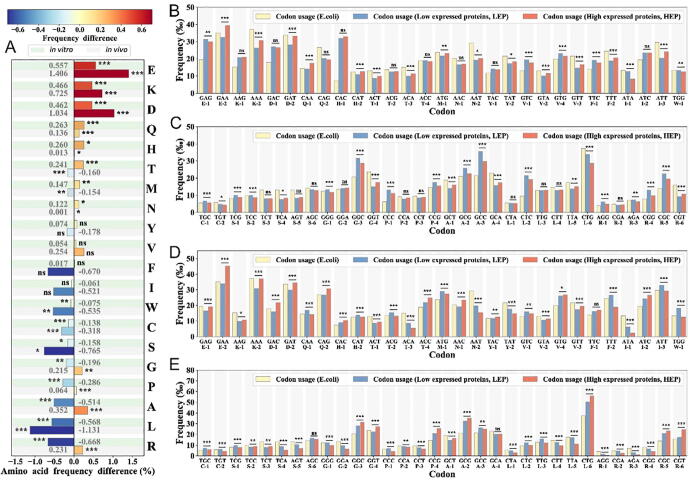
An initial analysis of the data revealed significantly different usage of amino acids between the low- and high-expression proteins (Fig. 2A). Among the eleven amino acids (E, K, D, Q, H, T, M, N, Y, V, and F) preferred in the highly expressed proteins (Fig. 2B), four were charged amino acids (E, K, D, and H) and eight amino acids (E, K, D, Q, H, N, Y, and F) were encoded by only two codons. Additionally, the amino acid E, K and D were also preferred in the endogenous proteins of E. coli (Fig. S3). However, among the other nine amino acids (R, L, A, P, G, S, C, W, and I) preferentially found in low-expression proteins (Fig. 2C), only one was a charged amino acid (R), and six of these amino acids (R, L, A, P, G, and S) were encoded by four or six codons.
Fig. 2.
Comparison of the amino acid or codon frequencies in lowly and highly expressed proteins. A. Amino acids usage differences between the lowly and highly expressed proteins. B-C. Codon frequencies in the highly and-lowly expressed proteins, and in the endogenous E. coli genome. D-E. Codon frequencies in the highly and-lowly expressed proteins, and in the exogenous E. coli genome. In addition, “ns” denotes no significant, “*” denotes 0.01 < p-value ≤ 0.05, “**” denotes 0.001 < p-value ≤ 0.01, and “***” denotes p-value ≤ 0.001.
In addition, five of the nine amino acids preferentially used in low-expression proteins (R, S, G, C and P) contained the codons AGG, AGT, GGT, TGT, and CCC which have been established to significantly affect in vivo translation speed in E. coli (Loss of attenuation >3) [31]. Here, the loss of attenuation of the codon was relative score that was the β-galactosidase activity of each codon construct divided by the activity of the wild-type construct that contains the CAC His codon [31]. The high loss of attenuation of the codon indicated that the codon could decrease the translation speed, and the vice versa [31]. Notably, in high-expression proteins, the codons encoded any of the preferred eleven amino acids that did not greatly affect the translation rate [31]. The average attenuation loss of preferential high-expression codons was 1.57 ± 0.53, significantly (p < 0.01) lower than the average attenuation loss (2.75 ± 2.12) of preferred codons in the low expression proteins [31] (Table S5). Therefore, the average translation rate of high expression proteins was significantly greater than that of the low expression proteins. Additionally, the correlation coefficients between the codon usage vector of the endogenous genes in the E. coli and the genes for the high or low expressed heterologous proteins (Fig. S3B and S3C) were 0.58 and 0.45, respectively. These results revealed that highly expressed proteins were more likely to use optimal synonymous codons than low-expression proteins, which could facilitate faster translation of those proteins. In light of these findings, we hypothesized that the selection of charged residues [32] and optimal synonymous (i.e., penalty-free) codons in highly expressed proteins likely enhanced their solubility and translation rate [33], [34], respectively.
3.3. Evaluation of deep learning model performance
For training the classifiers to accurately predict the effects of a given mutation on expression, we used the dataset constructed in the above section [17]. Since each codon in a given gene sequence could also be categorized according to synonymous codon number, i.e., number of codons encoding the same amino acid (1 to 6), the specific amino acid it called (1 to 20), or its specific nucleotide combination (1 to 61) (Table S1), three classifiers were constructed based on these three coding patterns, respectively. Receiver Operating Characteristics curve (ROC) and Precision-Recall curve (PRC) analyses were used to evaluate the predictive accuracy of each classifier through 10-fold cross validation. Notably, AUROC and AUPRC scores of the classifier that used specific nucleotide combinations were both markedly higher than those of the other two classification strategies, which were 0.764 and 0.751, respectively (Fig. 3B). Additionally, we also considered several alternative metrics, including the prediction, accuracy, F1 score, precision score, and recall score; all of these indexes supported classification based on the 61 combinations of nucleotides as the highest performing model for predicting protein expression levels (Fig. 3C). This result demonstrated the importance effect of codon usage on protein levels.
Fig. 3.
Evaluation predictive performance of the three constructed models. A-B. Receiver operator characteristic and precision recall curves for the three models output based on the results of 10-fold cross-validation. C. Model evaluation metrics. D. Prediction results of the three models on the independent test set class4 and class5. In addition, “ns” denotes no significant, “*” denotes 0.01 < p-value ≤ 0.05, and “**” denotes p-value ≤ 0.01.
When the three prediction models were applied to groups 4 and 5, the results showed that group 5 proteins had a significantly greater propensity for high expression in E. coli than that of proteins in the 4th group (p < 0.001) (Fig. 3D). The ratios of predicted average highly expressed propensity of the genes between groups 5 and 4 were 1.01, 1.09, and 1.13 for the synonymous codon number-, amino acid-, and nucleotide combination-based models, respectively. Thus, these collective results all indicated that specific nucleotide combinations were most informative aspect of codon frequency for predictive modeling of soluble protein expression. We therefore selected the model trained with specific nucleotide combination frequencies to design subsequent mutations.
3.4. Correlation between the predicted high-level expression propensity and protein abundance
To evaluate the relationship between the predicted high-level expression propensity and the protein abundance, we collected the protein abundance (PA) data of seven species of unicellular organisms from Paxdb and predicted the high-level expression propensity of those endogenous genes with the three coding schemes. As shown in Table 2 and Fig. S4, most of the spearman rank correlations of the predicted high-level propensity with the coding scheme of 61 combinations of nucleotides were higher than the other two coding schemes. Additionally, among all spearman rank correlations of species, the correlation between the PA of E. coli and the predicted high-level propensity with the coding scheme of 61 combinations of nucleotides exhibited the highest score which was 0.4581. The results indicated that the constructed models based on the analysis of heterologous gene expression propensity in the E. coli expression system gave significant results when predicted endogenous gene expression in various organisms across the tree of life. This suggest that there are universal gene expression codes that our model was able to detect.
Table 2.
Spearman rank correlation of the predicted high-level propensity with PA.a
| Number of Genes | r(Log(PA), Pre1)b | r(Log(PA), Pre2)c | r(Log(PA), Pre3)d | |
|---|---|---|---|---|
| Bacteria | ||||
| E. coli | 3063 | 0.1342 | 0.4036 | 0.4581 |
| S. enterica | 2200 | 0.0447 | 0.2240 | 0.2494 |
| B. subtilis | 2943 | 0.2130 | 0.3270 | 0.3718 |
| S. aureus | 1166 | 0.0443 | 0.2939 | 0.3816 |
| S. pyogenes | 1064 | 0.0531 | 0.2685 | 0.3290 |
| Archaea | ||||
| T. gammatolerans | 1092 | −0.1538 | 0.2637 | 0.1428 |
| Fungi | ||||
| S. cerevisiae | 4646 | −0.0582 | 0.3386 | 0.3617 |
Protein abundance data of genes from paxdb.
Pre1: Predicted high-level propensity with the coding scheme of the synonymous codon number.
Pre2: Predicted high-level propensity with the coding scheme of the specific amino acid.
Pre3: Predicted high-level propensity with the coding scheme of the specific nucleotide combination.
3.5. Mutation prediction to increase expression of laccase 13B22 and AtGDH
To further validate the MPEPE model, we selected two proteins, laccase 13B22 (GenBank number: MZ817083) and the glucose dehydrogenase, AtGDH (GenBank number: XM_001216916), both of which were reported to have high value in commercial applications and were not included in the training, validation and test dataset [35], [36]. Laccase 13B22 can be used in oxidative bioremediation of toxic xenobiotic compounds, while glucose dehydrogenase is an oxidoreductase that catalyzes the oxidation of glucose into gluconic acid δ-lactone. Although, the coding sequences of laccase 13B22 and glucose dehydrogenase FAD-AtGDH were both optimized for expression in E. coli, the two proteins still exhibited poor efficiency in their expression in E. coli, which we speculated was related to their codon composition.
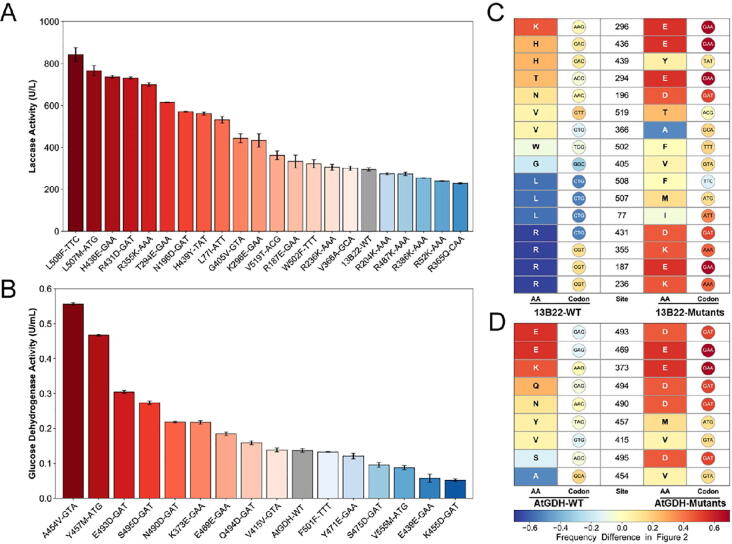
In order to ensure that both enzymes remained functional after modification for high, soluble expression, we first examined their conserved residues through protein sequence alignment and residue entropy analysis. Using BLASTP, we identified and downloaded sequences of 1335 and 2413 homologous proteins of 13B22 and FAD-AtGDH, respectively. Based on those collected sequences from NCBI, the position-specific amino-acid probability (PSAP) matrices for both enzymes were calculated. The PSAP cutoff value was set to 0.05, which was the average PSAP value of each amino acid at a given target residue (Figs. S5 and S6). The entropy of each residue was then determined and the higher the entropy, the lower the conservation at that site. For our purposes, we set the demarcation point to 0.65, and higher entropy values indicated that residues did not have conserved function at that position. Finally, the laccase 13B22 and FAD-AtGDH amino acid sequences were individually used as inputs for the MPEPE model to predict all single-point mutations that could improve their soluble expression in E. coli (see more details in the Methods section). As a result, there were 21 and 30 predicted mutations that were satisfied the above three conditions, located at the non-conserved sites and the predicted high-level expression propensity by the constructed model higher than that of the wild-type (Tables S5 and S6). Notably, most of these mutations were located at the C-terminal end and exposed at the protein surface (Fig. 4A and C). The AlphaFold 2.0 tool was used to simulate the three-dimensional structures of laccase 13B22 and FAD-AtGDH. The location of the mutation sites on the 3D structure were analyzed and their percent solvent accessibilities (PSA) were calculated using Discovery Studio (2016), respectively. The average PSA of the mutations of 13B22 and FAD-AtGDH were 43.67% and 32.12%, respectively (Fig. 4B and D, Table S6).
Fig. 4.
The entropy of the residue and the distribution of the mutations on the sequence and structure of laccase 13B22 and FAD-AtGDH. A–C. Residue entropy of the laccase 13B22 (A) and FAD-AtGDH (B). The black dot represents the location of the screened mutation. The strand, helix, and coil are the predicted secondary structure based on the method. B–D. The distribution of the mutations on the structure of laccase 13B22 and FAD-AtGDH.
3.6. Expression levels and enzyme activity validation of the predicted point mutations
To validate the effects of the predicted single point mutations on soluble protein expression in E. coli, we individually constructed the 21 and 30 single-point mutations into laccase 13B22 and FAD-AtGDH, respectively. The proteins of all of the variants was expressed in E. coli under the same induction and expression conditions. Since SDS-PAGE analysis could not clearly distinguish differences in expression between the 13B22 single point mutants and wild type (Fig. S7), we screened for laccase positive mutants using laccase activity assays with culture supernatant to characterize their soluble expression levels. By contrast, SDS-PAGE clearly indicated that FAD-AtGDH single point mutants were expressed at significantly higher levels than that of wild type (Fig. S8). The enzymatic activities of FAD-AtGDH variants and wild type in crude enzyme solution were also examined. The results showed that, among the 21 laccase 13B22 single point mutants, 16 mutants exhibited higher enzymatic activity than that of wild type. In particular, L508F-TTC showed the highest activity of 841.73 ± 33.25 U/L, 2.854 times higher than that of wild type (Fig. 5A, Table S7). Among the 30 FAD-AtGDH single point mutants, 9 mutants had higher enzymatic activity than the wild type, and of the 15 variants that retained activity after mutagenesis. A454V-GTA had the highest activity (555.49 ± 4.51 U/L), which was 3.05 times greater than that of the wild type (137.02 ± 4.92 U/L) (Fig. 5B, Table S8).
Fig. 5.
The detection of enzymatic activity and distribution of amino acid and codon of the mutants and wild-type of the laccase 13B22 and FAD-AtGDH. A. Measured enzymatic activity of the single-point mutants and wild-type of the laccase 13B22. B. The measured enzymatic activity of the single-point mutants and wild-type of the FAD-AtGDH. C-D. The amino acid and codon selection of the mutants and wild-type of the laccase 13B22(C) and FAD-AtGDH(D). The color bar represents the amino acid usage difference between the high- and low-level expressed genes.
Sequence analysis of these positive mutants revealed that glutamic acid (E) and lysine (K), encoded by two synonymous codons, appeared at higher frequency in the 13B22 and FAD-AtGDH variants, while arginine (R) or leucine (L), encoded by six synonymous codons, were less abundant in the variants than in the wild type (Fig. 5C and D). These findings suggested that most of variants with improved soluble expression likely benefitted from the charged amino acids and acceleration of translational rate in the protein C-terminal.
3.7. Characterization of variants with multiple point mutations
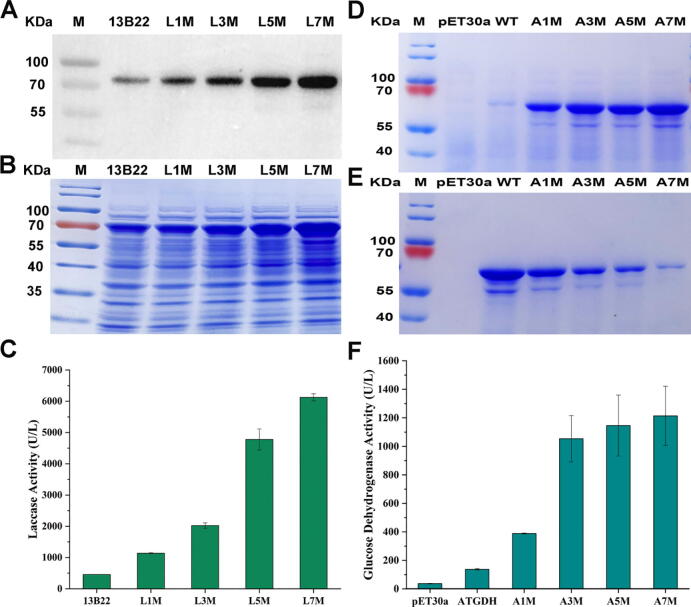
In order to determine whether combining the point mutations could further increase the soluble expression levels of the two proteins, we next generated variants harboring 3, 5, or 7 point mutations (selected based on the magnitude of their effects on soluble expression), and expressed them in E. coli. Soluble expression of 13B22 in culture supernatant gradually increased with increasing numbers of mutated sites (Fig. 6A and B). In particular, laccase mutant L7M (harboring L508F/L507M/H436E/R431D/R355K/T294E/N196D) showed the highest expression, approximately 3.49 ± 0.90 times that of wild type (Fig. 6A). In addition, laccase 13B22-L7M activity reached 3954.03 ± 74.03 U/L, approximately 12.40 times that of the wild type (294.938 ± 7.962 U/L) (Fig. 6C). The soluble protein expression of FAD-AtGDH by E. coli BL21 (DE3) also increased significantly with increasing numbers of mutation sites. Notably, AtGDH-A7M (carrying mutations A454V/Y457M/E493D/S495D/N490D/K373E/E469E) was apparently expressed almost entirely in the soluble fraction (Fig. 6D and E). The enzymatic activity of the AtGDH-A7M variant was 1213.48 ± 207.07 U/L, or 7.86 times greater than wild type (137.02 ± 4.92 U/L) (Fig. 6F). In summary, these results demonstrated that introducing mutations predicted by the preferential usage of charged amino acids in non-conserved sites and codons with fewer synonymous codons significantly improved the expression of these two proteins in E. coli, thereby validating the predictive accuracy of MPEPE.
Fig. 6.
Soluble expression and enzymatic activity assay of 13B22 and FAD-AtGDH. A. The Western-Blot of the expression of laccase 13B22 in supernatants in E. coli, L1M, L3M, L5M, and L7M represented 1, 3, 5, and 7 point mutants of laccase 13B22, respectively. B. The SDS-PAGE of the expression of laccase 13B22 in supernatants in E. coli, L1M, L3M, L5M, and L7M represented 1, 3, 5, and 7 point mutants of 13B22 respectively. C. The enzymatic activity of laccase 13B22, L1M, L3M, L5M, and L7M represented 1, 3, 5, and 7 point mutants of 13B22 respectively. D. The SDS-PAGE of the expression of FAD-AtGDH in supernatants in E. coli, A1M, A3M, A5M, and A7M represented 1, 3, 5, and 7 point mutants of FAD-AtGDH respectively. E. The SDS-PAGE of the expression of FAD-AtGDH in precipitations in E. coli, A1M, A3M, A5M, and A7M represented 1, 3, 5, and 7 point mutants of FAD-AtGDH respectively. F. The enzymatic activity of FAD-AtGDH, A1M, A3M, A5M, and A7M represented 1, 3, 5, and 7 point mutants of FAD-AtGDH respectively.
4. Discussion
In this study, we constructed DNN-based models for predicting mutations conducive to higher soluble protein expression in E. coli. In particular, we developed a predictive strategy (MPEPE) to screen for mutations that confer these properties and trained the model with gene sequence data calculated from publicly available, experimental, protein expression data. The dataset analysis showed the preferential use of four charged amino acids (E, K, D, and H) in the high-level expressed dataset. These results support previous studies that the charged amino acid can affect translation speed initiation and elongation [37], [38], [39].
In addition, we evaluated the high-level expression propensity of a human acetylcholinesterase (hAChE) variant bearing 51 mutations, which was approximately 2000-fold higher than that of the wild type in E. coli [40]. Based on our predictive scoring with MPEPE, the 51-point mutant of hAChE was 0.482, higher than the score of 0.463 for the wild type, which suggested an increased propensity for soluble expression. Moreover, the amino acids were mutated to the charged amino acids and the codons with high translational rate (Fig. S9).
Our results support the fact that expression levels are affected by the gene translation rate. In 2004, Wernisch and co-workers introduced the concept of tAI, which measures that adaptation of a coding region to the tRNA pool in the cell [41]; this measure was later generalized to fit various organism [42]. It was suggested in various additional studies that codons with higher adaptation to the tRNA pool (i.e. that are recognized by tRNAs with higher concentration in the cell) tend to have fast translational speed [43], [44]. For example, E. coli has four copies of the tRNA gene for codon GAA, encoding glutamate (E), and six copies of the tRNA gene for codon AAA, encoding lysine (K). By contrast, the E. coli genome contained one or zero copies of the vast majority of tRNA genes matching codons for arginine (R) and leucine (L), which both have several synonymous codons, coinciding with the rule we found through development of MPEPE [45]. In addition, the mutation sites that we found could positively affect 13B22 and FAD-AtGDH expression in this study were mainly concentrated in the C-terminus of the protein. These results were consistent with the findings reported in [41] (see also [46], [47]) which showed the N-terminus of proteins are translated at a slower rate than the downstream region, resulting in increased translation efficiency and ribosomal allocation.
The predictors designed here are based only on the coding sequence and where trained based on the analysis of the expression genes with the same promoter. Thus, the fact that there is a correlation of 0.46 between the predicted protein levels by our model and the protein abundance of endogenous genes in E. coli suggest that may suggest that at least 20% of the gene expression variability in E. coli can be explained by coding region features. This is a non-negligible value that should be considered when deigning heterologous genes and when studying genome evolution.
Compared to the relatively scattered data that might apply different expression vectors or different expression conditions used by Yu et al., which was only collected from the PDB database [14], [48], we used high-quality data published by Letso et al. strictly derived from E. coli under the same expression vector and conditions [17], to construct the MPEPE deep learning model. Although these experimental data cannot be clustered by traditional methods (Fig. S10), our model can effectively extract the features relevant to our protein engineering research question. In addition, our model was validated through mutagenesis and activity assays in laccase 13B22 and FAD-AtGDH, suggesting that the MPEPE provides highly accurate predictions, with broad potential applicability.
Data availability
The gene sequence of laccase 13B22 was uploaded to GenBank under the accession number MZ817083. The details of PCR primers are available in the Supplementary Data.
Funding
This work was supported by the National Key R&D Program of China [Grant No. 2021YFC2100300], the Central Public-interest Scientific Institution Basal Research Fund [grant numbers Y2019XK19, 1610392021008 and 1610392020001].
CRediT authorship contribution statement
Zundan Ding: Methodology, Validation, Data curation, Visualization, Writing – original draft. Feifei Guan: Writing – review & editing. Guoshun Xu: Visualization, Software, Writing – original draft. Yuchen Wang: Validation. Yaru Yan: Validation. Wei Zhang: Writing – review & editing. Ningfeng Wu: Writing – review & editing. Bin Yao: Supervision. Huoqing Huang: Conceptualization, Methodology. Tamir Tuller: Writing – review & editing. Jian Tian: Supervision, Project administration, Conceptualization, Methodology, Software, Data curation, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Prof. Haipeng Gong in Tsinghua University for help to construct the DNN models and Weitong Qin in Shanghai Jiaotong University for the analysis of the protein structure using AlphaFold 2.0 in this research. We also thank Yifan Wang in Biotechnology Research Institute, Chinese Academy of Agricultural Sciences for his suggestions and comments on the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.02.030.
Contributor Information
Zundan Ding, Email: dingzundan@caas.cn.
Feifei Guan, Email: guanfeifei@caas.cn.
Guoshun Xu, Email: xgs0114@126.com.
Yuchen Wang, Email: 13519669461@163.com.
Yaru Yan, Email: 18306429734@163.com.
Wei Zhang, Email: zhangwei02@caas.cn.
Ningfeng Wu, Email: wuningfeng@caas.cn.
Bin Yao, Email: binyao@caas.cn.
Huoqing Huang, Email: huanghuoqing@caas.cn.
Tamir Tuller, Email: tamirtul@tauex.tau.ac.il.
Jian Tian, Email: tianjian@caas.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Newton M.S., Arcus V.L., Gerth M.L., Patrick W.M. Enzyme evolution: innovation is easy, optimization is complicated. Curr Opin Struct Biol. 2018;48:110–116. doi: 10.1016/j.sbi.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 2.Zhao H., Xu Y.P., Li X.Y., Li G., Zhao H.F., Wang L.L. Expression and purification of a recombinant enterotoxin protein using different E. coli host strains and expression vectors. Protein J. 2021;40:245–254. doi: 10.1007/s10930-021-09973-w. [DOI] [PubMed] [Google Scholar]
- 3.Bhatwa A., Wang W.J., Hassan Y.I., Abraham N., Li X.Z., Zhou T. Challenges associated with the formation of recombinant protein inclusion bodies in Escherichia coli and strategies to address them for industrial applications. Front Bioeng Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.630551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen J.T., Fong J., Fong D., Fong T., Lucero R.M., Gallimore J.M., et al. Soluble expression of recombinant midgut zymogen (native propeptide) proteases from the Aedes aegypti Mosquito utilizing E-coli as a host. Bmc Biochem. 2018;19:12. doi: 10.1186/s12858-018-0101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azizi N., Shahpiri A. Functional characterization of Helianthus annuus phytochelatin synthase (HaPCS): Gene expression and protein profiles of HaPCS responding to arsenic and evaluation of arsenic accumulation in engineered bacteria expressing HaPCS. Environ Exp Bot. 2021;187 [Google Scholar]
- 6.Grzegorz, Kudla, Andrew, W., Murray, David, Tollervey, Joshua, B., Plotkin, Coding-sequence determinants of gene expression in Escherichia coli. Science 2009;324:255–58. [DOI] [PMC free article] [PubMed]
- 7.de Marco A. Protocol for preparing proteins with improved solubility by co-expressing with molecular chaperones in Escherichia coli. Nat Protoc. 2007;2:2632–2639. doi: 10.1038/nprot.2007.400. [DOI] [PubMed] [Google Scholar]
- 8.Deng Y. Glu659Leu substitution of recombinant HIV fusion inhibitor C52L induces soluble expression in Escherichia coli with equivalent anti-HIV potency. Protein Eng Des Sel. 2011;24:545–551. doi: 10.1093/protein/gzr013. [DOI] [PubMed] [Google Scholar]
- 9.Wang T., Badran A.H., Huang T.P., Liu D.R. Continuous directed evolution of proteins with improved soluble expression. Nat Chem Biol. 2018;14:972–980. doi: 10.1038/s41589-018-0121-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gil-Garcia, Marcos, Ba no-Polo, Manuel, Varejao, Nathalia, Jarnroz, Michal, Kuriata Aleksander, Combining structural aggregation propensity and stability predictions to redesign protein solubility. Mol Pharm, 2018;15:3846–59. [DOI] [PubMed]
- 11.Xia W., Zhang X., Gao Q., Gao X. Adversarial self-supervised clustering with cluster-specificity distribution. Neurocomputing. 2021;449:11. [Google Scholar]
- 12.Wilkinson D.L., Harrison R.G. Predicting the solubility of recombinant proteins in Escherichia coli. Nat Biotechnol. 1991;1258:403. doi: 10.1038/nbt0591-443. [DOI] [PubMed] [Google Scholar]
- 13.Diaz A.A., Tomba E., Lennarson R., Richard R., Bagajewicz M.J., Harrison R.G. Prediction of protein solubility in Escherichia coli using logistic regression. Biotechnol Bioeng. 2010;105:374–383. doi: 10.1002/bit.22537. [DOI] [PubMed] [Google Scholar]
- 14.Wu X., Yu L. EPSOL: sequence-based protein solubility prediction using multidimensional embedding. Bioinformatics. 2021;btab463 doi: 10.1093/bioinformatics/btab463. [DOI] [PubMed] [Google Scholar]
- 15.Fu H., Liang Y., Zhong X., Pan Z., Huang L., Zhang H., et al. Codon optimization with deep learning to enhance protein expression. Sci Rep. 2020;10:17617. doi: 10.1038/s41598-020-74091-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tunney R., McGlincy N.J., Graham M.E., Naddaf N., Pachter L., Lareau L.F. Accurate design of translational output by a neural network model of ribosome distribution. Nat Struct Mol Biol. 2018;25:577–582. doi: 10.1038/s41594-018-0080-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boel G., Letso R., Neely H., Price W.N., Wong K.H., Su M., et al. Codon influence on protein expression in E. coli correlates with mRNA levels. Nature. 2016;529:358. doi: 10.1038/nature16509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price W.N., 2nd, Handelman S.K., Everett J.K., Tong S.N., Bracic A., Luff J.D., et al. Large-scale experimental studies show unexpected amino acid effects on protein expression and solubility in vivo in E. coli. Microb Inform Exp. 2011;1:6. doi: 10.1186/2042-5783-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M., Herrmann C.J., Simonovic M., Szklarczyk D., von Mering C. Version 4.0 of PaxDb: protein abundance data, integrated across model organisms, tissues, and cell-lines. Proteomics. 2015;15:3163–3168. doi: 10.1002/pmic.201400441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rampasek L., Goldenberg A. TensorFlow: Biology's gateway to deep learning? Cell Syst. 2016;2:12–14. doi: 10.1016/j.cels.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 21.Meng X., Yang L., Liu H., Li Q., Xu G., Zhang Y., et al. Protein engineering of stable IsPETase for PET plastic degradation by Premuse. Int J Biol Macromol. 2021;180:667–676. doi: 10.1016/j.ijbiomac.2021.03.058. [DOI] [PubMed] [Google Scholar]
- 22.Tian J., Wu N., Guo X., Guo J., Zhang J., Fan Y. Predicting the phenotypic effects of non-synonymous single nucleotide polymorphisms based on support vector machines. BMC Bioinf. 2007;8:450. doi: 10.1186/1471-2105-8-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirsch R.D., Etienne J. An improved PCR-mutagenesis strategy for two-site mutagenesis or sequence swapping between related genes. Nucleic Acids Res. 1998;26:1848–1850. doi: 10.1093/nar/26.7.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrock J., Russel D.W. manual (3rd ed.) 2001. Molecular cloning: A laboratory. [Google Scholar]
- 26.Nasoohi N., Khajeh K., Mohammadian M., Ranjbar B. Enhancement of catalysis and functional expression of a bacterial laccase by single amino acid replacement. Int J Biol Macromol. 2013;60:56–61. doi: 10.1016/j.ijbiomac.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 27.Brander S., Mikkelsen J.D., Kepp K.P. Characterization of an alkali- and halide-resistant laccase expressed in E. coli: CotA from Bacillus clausii. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0099402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durão P., Chen Z., Fernandes A.T., Hildebrandt P., Murgi Da D.H., Todorovic S., et al. Copper incorporation into recombinant CotA laccase from Bacillus subtilis: characterization of fully copper loaded enzymes. J Biotechnol. 2008;13:183–193. doi: 10.1007/s00775-007-0312-0. [DOI] [PubMed] [Google Scholar]
- 29.Yue Q., Yang Y., Zhao J., Zhang L., Xu L., Chu X., et al. Identification of bacterial laccase cueO mutation from the metagenome of chemical plant sludge. Bioresour Bioprocess. 2017;4:48. [Google Scholar]
- 30.Yang Y., Huang L., Wang J., Xu Z. Expression, characterization and mutagenesis of an FAD-dependent glucose dehydrogenase from Aspergillus terreus. Enzyme Microb Technol. 2015;68:43–49. doi: 10.1016/j.enzmictec.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 31.Chevance F.F., Le Guyon S., Hughes K.T. The effects of codon context on in vivo translation speed. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Requiao R.D., Fernandes L., de Souza H.J.A., Rossetto S., Domitrovic T., Palhano F.L. Protein charge distribution in proteomes and its impact on translation. Plos Comput Biol. 2017;13 doi: 10.1371/journal.pcbi.1005549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frumkin I., Lajoie M.J., Gregg C.J., Hornung G., Church G.M., Pilpel Y. Codon usage of highly expressed genes affects proteome-wide translation efficiency. Proc Natl Acad Sci USA. 2018;115:E4940–E4949. doi: 10.1073/pnas.1719375115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bertalovitz A.C., Badhey M.L.O., McDonald T.V. Synonymous nucleotide modification of the KCNH2 gene affects both mRNA characteristics and translation of the encoded hERG ion channel. J Biol Chem. 2018;293:12120–12136. doi: 10.1074/jbc.RA118.001805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taghizadeh T., Talebian-Kiakalaieh A., Jahandar H., Amin M., Tarighi S., Faramarzi M.A. Biodegradation of bisphenol A by the immobilized laccase on some synthesized and modified forms of zeolite Y. J Hazard Mater. 2020;386 doi: 10.1016/j.jhazmat.2019.121950. [DOI] [PubMed] [Google Scholar]
- 36.Sakai G., Kojima K., Mori K., Oonishi Y., Sode K. Stabilization of fungi-derived recombinant FAD-dependent glucose dehydrogenase by introducing a disulfide bond. Biotechnol Lett. 2015;37:1091–1099. doi: 10.1007/s10529-015-1774-8. [DOI] [PubMed] [Google Scholar]
- 37.Tuller T., Veksler-Lublinsky I., Gazit N., Kupiec M., Ruppin E., Ziv-Ukelson M. Composite effects of gene determinants on the translation speed and density of ribosomes. Genome Biol. 2011;12:R110. doi: 10.1186/gb-2011-12-11-r110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trylska J., Konecny R., Tama F., Brooks C.L., 3rd, McCammon J.A. Ribosome motions modulate electrostatic properties. Biopolymers. 2004;74:423–431. doi: 10.1002/bip.20093. [DOI] [PubMed] [Google Scholar]
- 39.Dao Duc K., Song Y.S. The impact of ribosomal interference, codon usage, and exit tunnel interactions on translation elongation rate variation. PLoS Genet. 2018;14 doi: 10.1371/journal.pgen.1007166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldenzweig A., Goldsmith M., Hill S., Gertman O., Laurino P., Ashani Y., et al. Automated structure- and sequence-based design of proteins for high bacterial expression and stability. Mol Cell. 2016;63:337–346. doi: 10.1016/j.molcel.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuller T., Carmi A., Vestsigian K., Navon S., Dorfan Y., Zaborske J., et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 42.Sabi R., Volvovitch Daniel R., Tuller T. stAIcalc: tRNA adaptation index calculator based on species-specific weights. Bioinformatics. 2017;33:589–591. doi: 10.1093/bioinformatics/btw647. [DOI] [PubMed] [Google Scholar]
- 43.Tuller T., Waldman Y.Y., Kupiec M., Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci U S A. 2010;107:3645–3650. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Supek F., Smuc T. On relevance of codon usage to expression of synthetic and natural genes in Escherichia coli. Genetics. 2010;185:1129–1134. doi: 10.1534/genetics.110.115477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.dos Reis M., Savva R., Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fredrick K., Ibba M. How the sequence of a gene can tune its translation. Cell. 2010;141:227–229. doi: 10.1016/j.cell.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuller T., Zur H. Multiple roles of the coding sequence 5' end in gene expression regulation. Nucleic Acids Res. 2015;43:13–28. doi: 10.1093/nar/gku1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smialowski P., Doose G., Torkler P., Kaufmann S., Frishman D. PROSO II–a new method for protein solubility prediction. FEBS J. 2012;279:2192–2200. doi: 10.1111/j.1742-4658.2012.08603.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The gene sequence of laccase 13B22 was uploaded to GenBank under the accession number MZ817083. The details of PCR primers are available in the Supplementary Data.