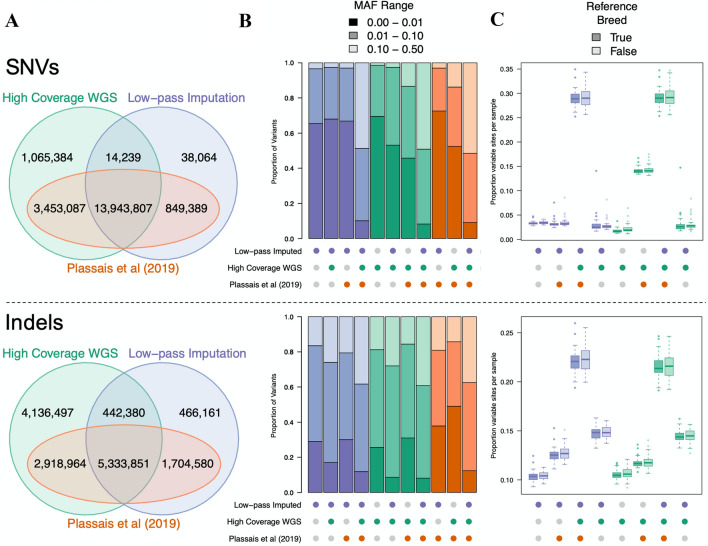
Fig. 2.
Genomic variant positions and their corresponding alleles are consistent across datasets. a Venn diagrams for SNVs and indels showing variants unique and shared across datasets. Datasets include the high-coverage WGS variant sites and low-pass imputed variant sites found across the 97 test samples and variants discovered in Plassais et al. (2019). Variants were identified as shared across datasets if the variant position, reference allele, and alternate allele were identical. b MAF distribution of each variant group from A. Variant groups are indicated by colored circles beneath the bar chart. Groups contain variants which are the intersect between the colored circles and do not contain variants found in the datasets represented by the gray circles. The color of each bar indicates the dataset used to calculate the MAF distribution and the shading level indicates the relevant MAF range. c Sites per sample in each variant group, where variant groups are presented as in B. Sites per sample are measured as the proportion of total sites within the relevant variant group that contain a non-reference allele for a particular sample. Samples have also been divided into two groups based on whether the respective breed also belongs to the Plassais et al. (2019) dataset and is, therefore, likely used in the imputation reference panel