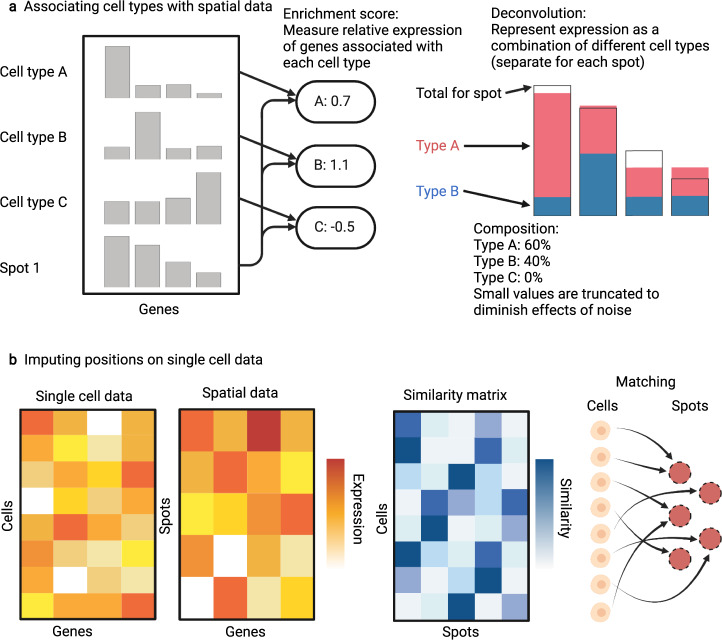
Fig. 4. Enhancing spatial transcriptomics data with scRNA-seq data.
This analysis step augments ST data using scRNA-seq data. a By using scRNA-seq data onto the spatial dataset, the composition of individual spots can be understood in terms of single cells, such as by computing enrichment scores, which measure the expression of certain gene sets (such as marker genes from a particular cell type) relative to the norm, or through deconvolution, which decomposes the overall expression data from a spatial spot into a combination of contributions from several cell types. b scRNA-seq data can be used to increase resolution of multi-cell ST data, by mapping cells to spatial locations, producing a spatial dataset at single-cell resolution. The primary choices in such methods are the computation of similarity scores between cells and spots, and the method by which matching is computed from the similarity matrix.