Abstract
The phylogenetic classification of Escherichia coli isolates is of great importance not only for understanding the populations of E. coli but also for clarifying the relationship between strains and diseases. The present study aimed to evaluate the prevalence of phylogenetic groups, antibiotic susceptibility pattern, and virulence genes among uropathogenic E. coli (UPEC) isolated from different parts of Iran through a systematic review and meta-analysis. Several international electronic sources, including Web of Science, PubMed, Scopus, and Embase, were searched (2000–2020) in order to identify the studies compatible with our inclusion criteria. The meta-analysis was performed using the metaprop program in the STATA (version 11) software. Based on our comprehensive search, 28 studies meeting the eligibility criteria were included in the meta-analysis. The pooled prevalence of phylogroups B2, D, B1, and A was 39%, 26%, 18%, and 8%, respectively. In addition, there was a significant heterogeneity among different phylogroups. However, according to the results of Begg’s and Egger’s tests, there were no significant publication bias in phylogroups B2, D, B1, and A. This research provided the first comprehensive study on phylogroups of UPEC isolated in Iran. Our findings indicated that phylogroup B2 and group D were the most predominant phylogenetic groups among UPEC isolates in various regions of Iran. In addition, we observed that certain phylogenetic groups are more antibiotic resistant than the others. It was also observed that the dissemination of virulent phylogroup B2 and D should be controlled via comprehensive infection control measures. Additionally, certain strategies should be developed for monitoring the antibiotic therapy.
Keywords: uropathogenic Escherichia coli , polygenetic groups, antibiotic resistant, virulence factor, meta-analysis
Introduction
Urinary tract infections (UTI), as one of the most prevalent pathological disorders, are the most problematic infectious diseases worldwide in both community and hospital settings (Terlizzi et al., 2017). UTI patients could be categorized into symptomatic and asymptomatic cases. Symptomatic UTI patients can be classified based on the degree of the severity into three classes, namely pyelonephritis (upper UTI, with kidney infection), cystitis (lower UTI, bladder infection), and urosepsis (Foxman, 2014; Smelov et al., 2016). Approximately 11 million individuals with UTI in the USA are annually referred to health centers, among whom 470,000 are hospitalized, incurring about $6 billion annually. It is known that Uropathogenic Escherichia coli (UPEC) is the main factor causing UTIs (Navidinia et al., 2018). The primary cause of UTIs is UPEC, both in community and hospital settings, which leads to a considerable rate of global mortality and morbidity (Tabasi et al., 2015). Antibiotic therapy is the only treatment for UTI; however, global spread of MDR bacterial strains has become a public health threat and major concern, particularly in patients with recurrent UTIs (Halaji et al., 2020; Sadeghi et al., 2020). Excessive use of a broad spectrum of antibiotics, such as fluoroquinolones, cephalosporins, and aminoglycosides, raise the cost of treatment and hospitalization (Fayyazi et al., 2020).
The major etiological agents associated with UTI are UPECs with the ability to successfully colonize the urinary tract. UPEC pathogenesis during UTIs occurs in an ascending manner as the following order: from the urethra to bladder to ureter to kidney to bloodstream (Terlizzi et al., 2017). A broad range of virulence factors and specific genes are expressed in UPEC isolates (Sadeghi et al., 2020). In fact, certain host cell types, including the stratified layers of bladder urothelium, such as differentiated superficial facet cells, less mature intermediate cells, and basal epithelial cells, can be breached by UPEC. Host cell invasion could facilitate the establishment and permanence of UPEC within the urinary tract (Lewis et al., 2016; Raeispour and Ranjbar, 2018).
In order to classify E. coli strains into one of the major phylogenetic classes of A, B1, B2, or D, a rapid and easy phylogenetic grouping technique based on triplex PCR has been developed to detect the genes chuA, yjaA, and TspE4. To improve the accuracy of their system, an extra gene target, arpA, has been added. This new quadruplex PCR is able to correctly assign E. coli strains in eight phylogroups, namely A, B1, B2, C, D, E, F, and one Escherichia cryptic clade I (Clermont et al., 2013; Najafi et al., 2018; Caméléna et al., 2019). The majority of strains responsible for extraintestinal infections belong to group B2 or to a lesser extent, to group D, while intestinal pathogenic and commensal isolates are observed in A and B1 (Molina-López et al., 2011; Lee et al., 2016). The phylogenetic classification of E. coli isolates is of great importance not only for understanding the populations of E. coli but also for clarifying the relationship between strains and diseases. E. coli sequence type 131 (ST131) is considered an important emerging pathogen among B2 strains, harboring multiple genes for resistance and virulence factors (VFs). The strains belonging to this group are mediated by the production of extended spectrum β-lactamases (ESBLs) and are resistant against most β-lactam antibiotics (Cristea et al., 2019). Expression of various markers of genetic virulence helps UPEC to cause infection within the urinary tract of the host (Picard et al., 1999; Johnson et al., 2001; Cristea et al., 2019). To the best of our knowledge, there is no available comprehensive information on the prevalence of phylogenetic groups and the related antibiotic susceptibility pattern and virulence genes among Iranian patients. Thus, the present study was conducted to determine the prevalence and distribution of phylogenetic groups, antibiotic susceptibility pattern, and virulence genes among UPEC isolated from different parts of Iran via a systematic review and meta-analysis.
Material and Methods
Search Strategies
The current study was carried out according to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines ( Supplementary Data ). A systematic literature search was conducted in the Web of Science, PubMed, Scopus, and Embase electronic databases. The search was limited to the articles published by Iranian authors from the beginning to the end of November 2020. The following terms, “Escherichia coli” OR “E. coli” OR “UPEC” OR “uropathogenic E. coli” OR “uropathogenic Escherichia coli” AND “phylogenetic group” OR “phylogroups” OR “phylogroup” OR “phylotypes “OR “phylogroups” AND “IRAN,” were searched as scientific keywords and phrases in the present survey.
Inclusion and Exclusion Criteria
To determine the articles meeting the inclusion criteria and reduce the risk of error, two reviewers screened independently the databases with the related keywords and reviewed the titles, abstracts, and full texts, and any discrepancies were resolved by consensus. The articles with the following criteria were included in the study: (1) cross-sectional, retrospective, and cohort studies indexed in the Web of Science or PubMed or Scopus database and reporting the prevalence of phylogenetic groups in E. coli isolates collected from the urine of patients with UTI and (2) those published worldwide with available English abstracts. Review articles, meta-analysis, or systematic articles, editorials, case report studies, letters to the editors, congress and meeting abstracts, studies where the sample size contains less than 10 isolates, studies with samples from environmental or nonclinical sources, articles without full text, duplicate publications, and articles with unclear and missing data were excluded.
Quality Assessment and Data Extraction
Five eligibility and quality assessment criteria were retrieved based on the Joanna Briggs Institute guidelines, and any disagreements were resolved by consensus. The following data were extracted for eligible studies: authors’ names, publication year, performed time, study location, characterization of the studied population, sample size, prevalence of phylogroups, virulence factor, and antibiotic resistance pattern.
Statistical Analysis
Analysis of data was carried out using the metaprop program in STATA statistical software, version 11.0 (Stata, College Station, TX, USA) (Nyaga et al., 2014). The pooled prevalence of phylogenic groups and associated antibiotic resistance and virulence factor with 95% confidence intervals (95% CIs) were estimated through the random effects model. In this meta-analysis, the CIs for proportions were computed using the score method. Statistical heterogeneity between the studies was calculated utilizing the Cochran Q Chi-square test and Cochrane I 2. The funnel plot, Begg’s rank correlation test, and Egger’s weighted regression tests were conducted to evaluate possible publication bias and any asymmetry appearing in the funnel plot, or p < 0.05 in the test was indicative of statistically significant publication bias (Begg, 1985). Possible sources of heterogeneity were calculated employing meta-regression analysis, and the subgroup analysis was performed based on the location of the study (region) and the types of patients (Zeng et al., 2015). Moreover, the sensitivity analysis was assessed with influence analysis and ignoring each study, followed by evaluating the estimated pooled prevalence in the absence of the excluded studies. In addition, the confounding effect of possible confounders, such as the time of the study (performed years), was evaluated by conducting meta-regression analyses.
Results
Database Search and Characterization of Studies
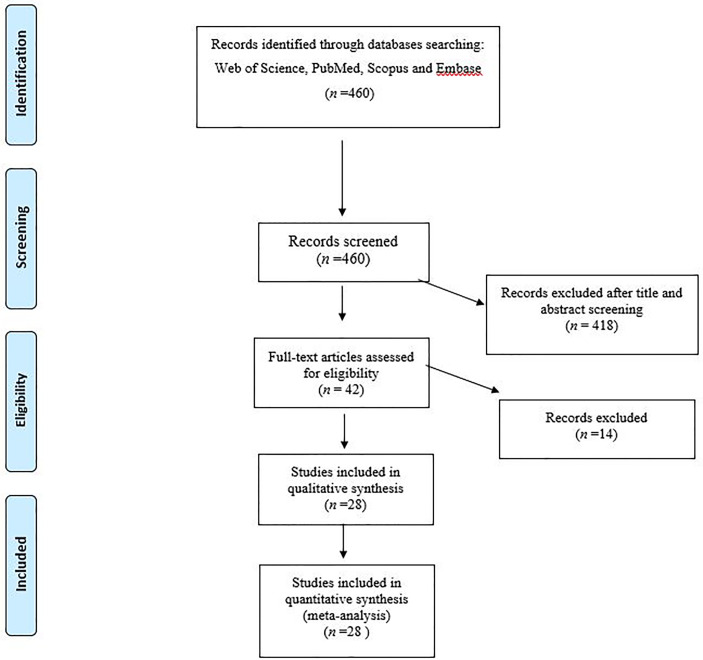
Based on our comprehensive search, 28 studies with eligibility criteria ( Figure 1 ) were included in the meta-analysis (Ramos et al., 2011; Navidinia et al., 2013; Adib et al., 2014; Alizade et al., 2014a; Alizade et al., 2014b; Hemati et al., 2014; Kazemnia et al., 2014; Derakhshandeh et al., 2015; Iranpour et al., 2015; Rahdar et al., 2015; Salmani et al., 2016; Sohrabi and Zeighami, 2016; Hashemizadeh et al., 2017; Hojabri et al., 2017; Salehzadeh and Zamani, 2018; Bahadori et al., 2019; Farajzadah Sheikh et al., 2019; Norouzian et al., 2019; Shahin et al., 2019; Staji et al., 2019; Bakhtiari et al., 2020; Moez et al., 2020; Ranjbar et al., 2020; Yazdanpour et al., 2020). Of the 28 included studies, 17 and 9 studies reported the prevalence of phylogenic groups from hospitalized and community patients, respectively. Also, two analyses were performed on both groups of patients. The full characteristics of the included studies are listed in Table 1 .
Figure 1.
Flow chart of the study selection for inclusion in the systematic review.
Table 1.
The main characteristics of studies included in the meta-analysis.
| Author | Published time | City | Region | Population studies | Number of E. coli isolates | A | B1 | B2 | D | ST131 | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ramos et al. | 2011 | Tehran | North | Hospitalized | 48 | – | – | 21 | – | – | Ramos et al. (2011) |
| Navidinia et al. | 2013 | Tehran | North | Hospitalized | 50 | 4 | 2 | 27 | 17 | – | Navidinia et al. (2013) |
| Adib et al. | 2014 | Kerman | Southeast | Hospitalized | 137 | 63 | 18 | 27 | 29 | – | Adib et al. (2014) |
| Alizade et al. | 2014 | Kerman | Southeast | Hospitalized | 45 | 4 | 7 | 19 | 15 | – | Alizade et al. (2014a) |
| Kazemnia et al. | 2014 | Urmia | North of West | Hospitalized | 25 | 8 | 0 | 10 | 7 | – | Kazemnia et al. (2014) |
| Alizadeh et al. | 2014 | Kerman | Southeast | Community | 122 | 55 | 5 | 9 | 53 | – | Alizade et al. (2014b) |
| Hemmati et al. | 2014 | Kerman | Southeast | Community | 92 | 23 | 8 | 29 | 32 | – | Hemati et al. (2014) |
| Iranpour and Najafi et al. | 2015 | Bushehr | South | Hospitalized | 140 | 1 | 7 | 55 | 4 | – | Iranpour et al. (2015) |
| Derakhshandeh et al. | 2015 | Shiraz | South | Hospitalized | 85 | 56 | – | 15 | 14 | – | Derakhshandeh et al. (2015) |
| Rahdar et al. | 2015 | Zabol | Southeast | Community | 100 | 17 | 6 | 55 | 22 | – | Rahdar et al. (2015) |
| Sohrabi et al. | 2016 | Zanjan | West | Hospitalized | 137 | 16 | – | 92 | 29 | – | Sohrabi and Zeighami (2016) |
| Salmani et al. | 2016 | Tehran Sanandaj | – | Hospitalized | 104 | 32 | 26 | 26 | 20 | – | Salmani et al. (2016) |
| Hashemizadeh et al. | 2017 | Kerman | Southeast | Hospitalized and community | 251 | 35 | 31 | 99 | 86 | – | Hashemizadeh et al. (2017) |
| Hashemizadeh et al. | 2017 | Kerman | Southeast | Hospitalized | 100 | 21 | 15 | 34 | 30 | 4 (B2) | Hashemizadeh et al. (2017) |
| 2 (A) | |||||||||||
| Hashemizadeh et al. | 2017 | Kerman | Southeast | Community | 151 | 14 | 16 | 65 | 56 | – | Hashemizadeh et al. (2017) |
| Salehzadeh et al. | 2017 | Rasht | North | Community | 100 | 14 | 6 | 52 | 28 | – | Salehzadeh and Zamani (2018) |
| Hojabri et al. | 2017 | Semnan | North | Hospitalized | 339 | – | – | 136 | 33 | 62 (B2) | Hojabri et al. (2017) |
| 1 (F) | |||||||||||
| Staji et al. | 2019 | Semnan | North | Community | 160 | 25 | 12 | 75 | 48 | – | Staji et al. (2019) |
| Yazdanpour et al. | 2020 | Zabol | Southeast | Community | 248 | 16 | 12 | 167 | 53 | – | Yazdanpour et al. (2020) |
| Farajzadah Sheikh et al. | 2019 | Ahvaz | Southwest | Hospitalized and community | 232 | 37 | 18 | 42 | 135 | – | Farajzadah Sheikh et al. (2019) |
| Farajzadah Sheikh et al. | 2019 | Ahvaz | Southwest | Hospitalized | 139 | 19 | 9 | 23 | 90 | – | Farajzadah Sheikh et al. (2019) |
| Farajzadah Sheikh et al. | 2019 | Ahvaz | Southwest | Community | 93 | 18 | 9 | 19 | 45 | – | Farajzadah Sheikh et al. (2019) |
| Bahadori et al. | 2018 | Shiraz | South | Community | 90 | 13 | 6 | 53 | 18 | – | Bahadori et al. (2019) |
| Norouzian et al. | 2019 | Tehran | North | Hospitalized | 106 | 22 | 10 | 52 | 22 | – | Norouzian et al. (2019) |
| Najar Peerayeh et al. | 2019 | Tehran | North | Hospitalized | 16 | 3 | 2 | 7 | 4 | – | Shahin et al. (2019) |
| Morovati et al. | 2020 | Hamadan | West | Hospitalized | 140 | 23 | 8 | 39 | 4 | – | Moez et al. (2020) |
| Ranjbar et al. | 2020 | Tehran | North | Hospitalized | 60 | 1 | 3 | 50 | 6 | – | Ranjbar et al. (2020) |
| Bakhtiari et al. | 2020 | Hamadan | West | Hospitalized | 113 | 23 | 5 | 50 | 35 | – | Bakhtiari et al. (2020) |
Prevalence of Phylogroup B2
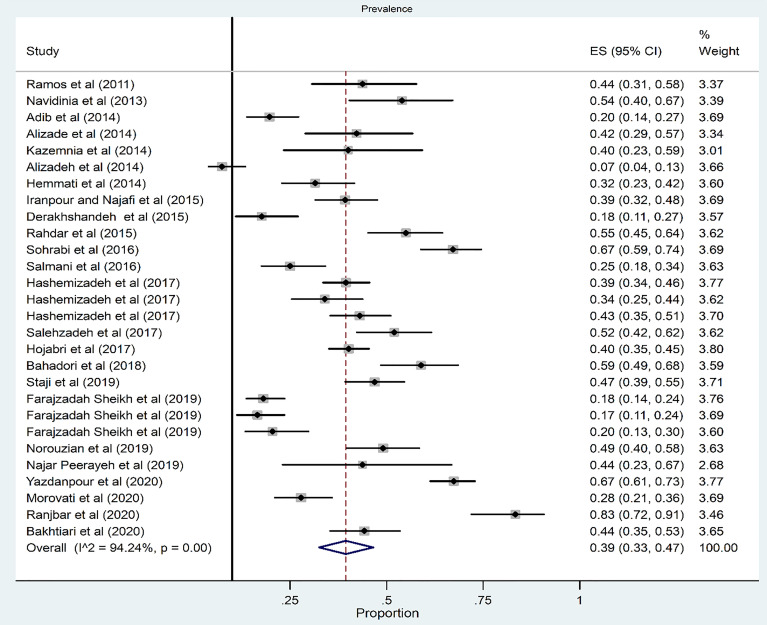
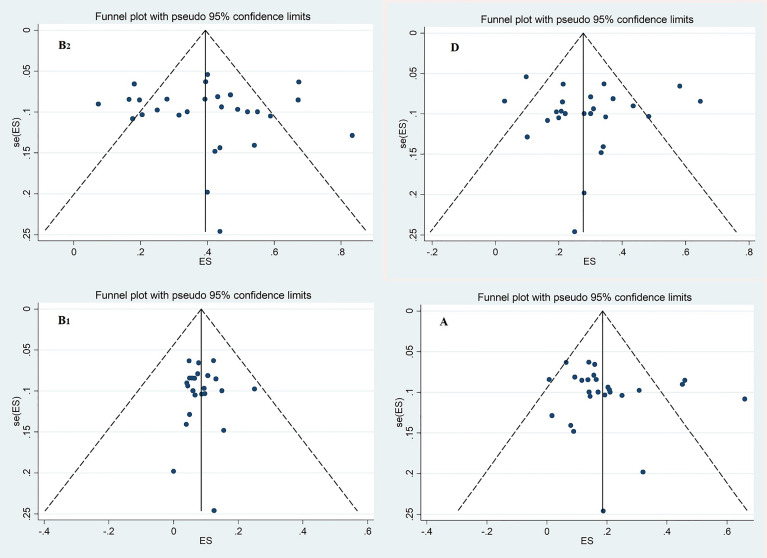
The pooled prevalence of phylogroup B2 among 28 studies was 39% (95% CI: 33–47) ( Figure 2 ). There was a significant heterogeneity among the 28 studies (χ 2 = 468.47; p < 0.001; I 2 = 94.24%). The funnel plot for publication bias did not show any evidence of asymmetry ( Figure 3 ). According to the results of Begg’s (Z = 0.51, p = 0.60) and Egger’s tests (t = 0.27, p = 0.79), there was no significant publication bias ( Figure 3 ).
Figure 2.
Forest plot of the meta-analysis of phylogroup B2 prevalence among UPEC isolates.
Figure 3.
Funnel plot for evaluation of publication bias; Phylogenetic groups B2, D, B1 and A.
The subgroup analysis results based on region indicated that the highest and lowest overall occurrence of phylogroup B2 was 52% and 18% in the north and southwest regions, respectively ( Supplementary Figure 1 and Table 1 ). The subgroup analysis results based on source of patients indicated that the overall prevalence of phylogroup B2 in the hospitalized and community patients was 40% and 41%, respectively ( Supplementary Figure 2 and Table 1 ).
Prevalence of Phylogroup D
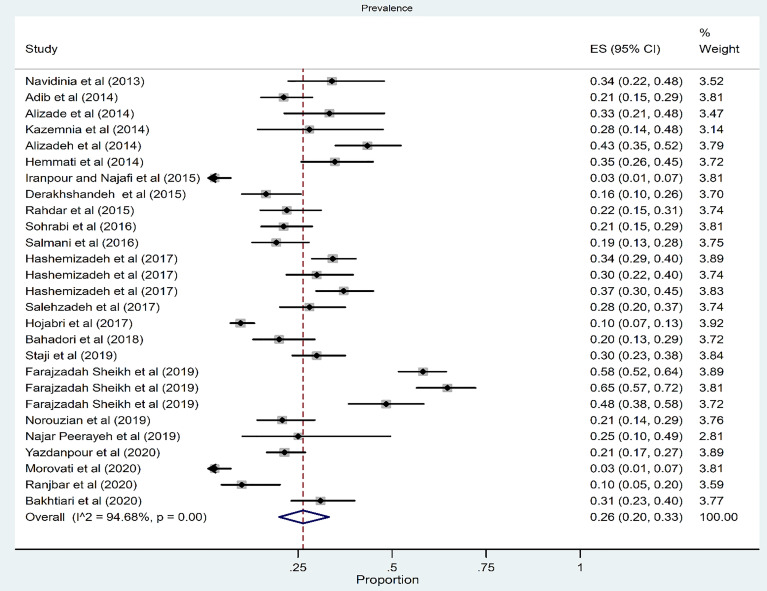
Among 27 studies, the pooled prevalence of phylogroups D was 26% (95% CI: 20–33) ( Figure 4 ). There was a significant heterogeneity for phylogroup D among the 27 studies (χ 2 = 488.47; p < 0.001; I 2 = 94.68%). The funnel plot for publication bias in the four phylogroups did not show any evidence of asymmetry ( Figure 3 ). Accordingly, the results of Begg’s (Z = 0.1, p = 0.91) and Egger’s tests (t = 0.09, p = 0.93) showed no significant publication bias in phylogroup D ( Figure 3 ).
Figure 4.
Forest plot of the meta-analysis of phylogroup D prevalence among UPEC isolates.
The subgroup analysis results based on region indicated that the highest and lowest overall prevalence of phylogroup D was in the southwest (58%) and south (12%) regions, respectively ( Supplementary Figure 3 and Table 2 ).
The subgroup analysis results based on source of patients indicated that the overall prevalence of phylogroup D was 21% in hospitalized and 31% in community patients ( Supplementary Figure 4 and Table 2 ).
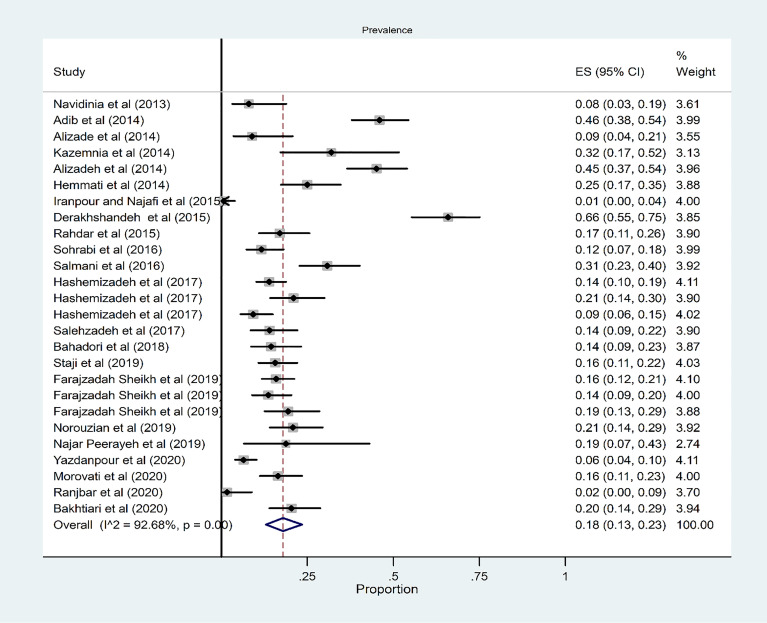
Prevalence of Phylogroup A
The pooled prevalence of phylogroup A among 26 studies was 18% (95% CI: 13–23) ( Figure 5 ). There was a significant heterogeneity among the 26 studies (χ 2 = 341.53; p < 0.001; I 2 = 92.68%). The funnel plot for publication bias in the four phylogroups did not show any evidence of asymmetry ( Figure 3 ). According to the results of Begg’s (Z = 1.52, p = 0.36) and Egger’s tests (t = 0.93, p = 0.36), there was no significant publication bias ( Figure 3 ). The subgroup analysis results based on region indicated that the highest and lowest overall occurrence of group A was in the northwest (32%) and north (12%) regions, respectively ( Supplementary Figure 5 and Table 3 ). The subgroup analysis results based on source of patients indicated that the overall prevalence of phylogroup A in the hospitalized and community patients were 19% and 17%, respectively ( Supplementary Figure 6 and Table 3 ).
Figure 5.
Forest plot of the meta-analysis of phylogroup A prevalence among UPEC isolates.
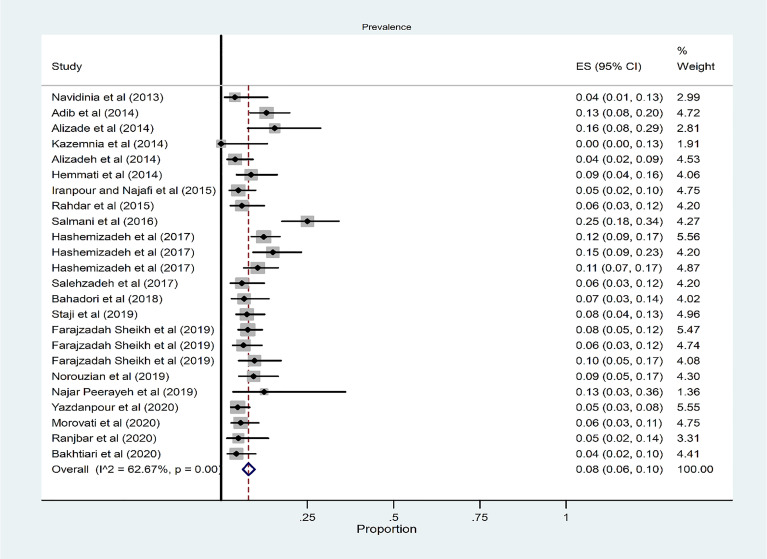
Prevalence of Phylogroup B1
From 24 studies, the pooled prevalence of phylogroup B1 was 8% (95% CI: 6–10) ( Figure 6 ). There was a significant heterogeneity for phylogroup B1 among the 24 studies (χ 2 = 61.62; p < 0.001; I 2 = 62.67%). According to the results of Begg’s (Z = 0.07, p = 0.94) and Egger’s tests (t = 0.06, p = 0.95) for phylogroup B1, there was no significant publication bias ( Figure 3 ).
Figure 6.
Forest plot of the meta-analysis of phylogroup B1 prevalence among UPEC isolates.
The subgroup analysis results based on region indicated that the highest and lowest overall occurrence of phylogroup B1 was 10% in the west and 0% in the south region ( Supplementary Figure 7 and Table 4 ). Also, the overall prevalence of phylogroup B1 was 8% and 7% in the hospitalized and community patients, respectively ( Supplementary Figure 8 and Table 4 ).
Sensitivity Analysis and Meta-Regression
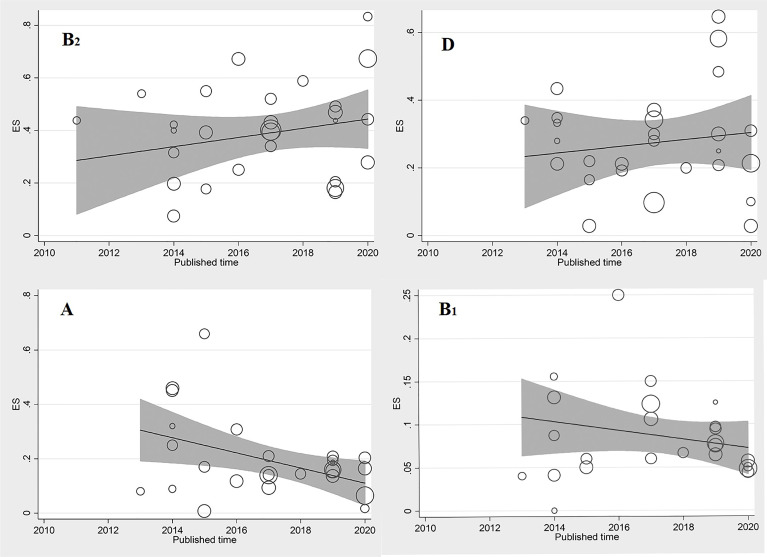
Meta-regression results indicated that the prevalence of phylogroups B2, D, and B1 among UPEC isolates was not significantly associated with year, coefficients: 0.01442 (95% CI: −0.01533–0.04418, p = 0.32), 0.005076 (95% CI: −0.025178–0.03533, p = 0.73), and −0.00513 (95% CI: −0.02311–0.01283, p = 0.55), respectively. Furthermore, no significant increasing trend was observed over time on the estimated pooled prevalence of phylogroups B2, D, and B1 in the included studies ( Figure 7 ).
Figure 7.
Meta regression of the log-event rates by year (phylogroups B2, D, A, and B1).
Meta-regression results indicated that the prevalence of phylogroup A among UPEC isolates was significantly associated with year, coefficients: 0.026443 (95% CI: −0.05079–0.00209, p = 0.034). Furthermore, significant decreasing trend was observed over time on the estimated pooled prevalence of phylogroup A in the included studies ( Figure 7 ).
Moreover, the results of the influence analysis demonstrating that none of the studies affect the estimated pooled prevalence of phylogroups B2, D, A, and B1. In addition, we excluded each study and examined the outcome. The sensitivity analyses also showed no significant change in the estimated pooled prevalence in the meta-analysis results after excluding studies with relatively lower quality [ Supplementary Figure 9 (A: B2, B: D, C: A, D: B1)].
Prevalence of Antibiotic Resistance Among Phylogroups
According to the antibiotic resistance pattern in phylogroup B2, ampicillin had the highest antibiotic resistance rate at 76% (95% CI: 58–90; I 2 = 89.57%; n = 6 studies), followed by cefixime at 70% (95% CI: 34–96; I 2 = 90.84%; n = 3 studies), while nitrofurantoin and imipenem had the lowest resistance rates at 2% (95% CI: 0–5; I 2 = 35.65%; n = 5 studies) and 7% (95% CI: 0–21; I 2 = 92.52%; n = 6 studies), respectively ( Supplementary Table 5 ).
In phylogroup D, the highest antibiotic resistance rate was against cefixime with 80% (95% CI: 53–98; I 2 = 45.54%; n = 3 studies), followed by ampicillin with 64% (95% CI: 28–94; I 2 = 93.87%; n = 6 studies), while amikacin 5% (95% CI: 0–15; I 2 = 60.68%; n = 5 studies) and ceftriaxone 17% (95% CI: 0–54; I 2 = 78.02%; n = 4 studies) had the lowest antibiotic resistance rate. Also, in this group, there was no resistance to nitrofurantoin ( Supplementary Table 6 ).
In group A, ampicillin had the highest antibiotic resistance rate at 59% (95% CI: 27–89; I 2 = 75.65%; n = 6 studies), followed by nalidixic acid at 58% (95% CI: 6–100; I 2 = 93.45%; n = 6 studies), while imipenem and amikacin were the most effective antibiotics with resistance rates of 1% (95% CI: 0–12; I 2 = 61.83%; n = 6 studies) and 3% (95% CI: 0–16; I 2 = 58.77%; n = 5 studies), respectively. Also, there was no resistance to nitrofurantoin in this group ( Supplementary Table 7 ).
Moreover, the highest antibiotic resistance rate in group B1 was against ampicillin with 32% (95% CI: 0–90; I 2 = 3.01%; n = 6 studies), followed by ciprofloxacin with 30% (95% CI: 16–46; I 2 = 0%; n = 5 studies), while nalidixic acid 1% (95% CI: 0–56; I 2 = 53.96%; n = 6 studies) and gentamicin 2% (95% CI: 0–10; I 2 = 27.07%; n = 6 studies) had the lowest antibiotic resistance rates. Also, in this group, there was no resistance to imipenem, cefotaxime, amikacin, nitrofurantoin, and ceftazidime ( Supplementary Table 8 ).
Prevalence of Virulence Factor Among Phylogroups
The results of the investigation of virulence factors into phylogenetic groups indicated that the highest prevalence among all groups was related to gene fimH, followed by gene papC.
In group B2, afa and hly genes with 7% and 21% frequency had the lowest prevalence among virulence factors ( Supplementary Table 9 ). In group D, the lowest prevalence of virulence factors was related to gene hly with 10% and papEF with 11% ( Supplementary Table 10 ). Also, in group A, the lowest prevalence of virulence factors was related to gene afa with 1% and cnfi with 1% ( Supplementary Table 11 ). Finally, papEF and hly genes with 4% and 5% prevalence were the lowest prevalence in group B1 ( Supplementary Table 12 ).
Prevalence of ST131 Among Phylogroups
Our finding revealed that among all of the studies included, only two reported ST131 among different phylogenetic groups. In total, sixty-nine isolates were ST131; of these isolates, 66, 2, and 1 belong to phylogenetic groups B2, A, and F, respectively.
Discussion
According to the phylogenetic background, E. coli strains showed moderate levels of recombination in the species (Tenaillon et al., 2010; Dixit et al., 2015; Touchon et al., 2020) and a strong phylogenetic structure with eight main phylogroups, four of which (A, B1, B2, and D) showed the majority of the strains and four others (C, E, F, and G) are more scarce. Moreover, these phylogroups apparently differ in their phenotypic and genotypic characteristics within and across phylogroups, such as their antibiotic-resistance profiles and their growth rate (Touchon et al., 2020).
Monitoring and evaluating the E. coli genotypic characteristics from urine resources provide useful data on the epidemiology of diseases in various geographical areas (Momtaz et al., 2013; Ranjbar and Farahani, 2018). To the best of our knowledge, the present work is the first comprehensive meta-analysis investigating the occurrence of phylogroup classes in UPEC isolates. We also investigated into the occurrence of phylogroup in different geographical locations and based on the type of patients.
Phylogenetic analysis indicated that the majority of UPEC isolates belonged to phylogroup B2 (39%), followed by group D (26%), group A, and group B1, which is in agreement with the study conducted by Munkhdelger et al. (2017), where B2 (33.8%) was the dominant phylogroup followed by D (28.4%), A (19.6%), and B1 (18.2%).
The majority of the studies on the phylogenetic grouping among UPEC have reported a similar distribution, such as studies conducted in China (Zhao et al., 2015), South Korea (Lee et al., 2016), Denmark (Ejrnæs et al., 2011), Pakistan (Bashir et al., 2012), Ethiopia (Dadi et al., 2020), Mexico (Paniagua-Contreras et al., 2017), and France (Dubois et al., 2010), in which it was found that the majority of isolates of E. coli predominantly belong to phylogenetic group B2. In this systematic review, we noted a high prevalence of phylogroup B2 in UPEC isolates, reflecting the importance of investigating and addressing the prevalence of isolates belonging to this group since they revealed a partially high level of antibiotic resistance and virulence factors.
Commensal populations of E. coli include stable genetic isolates with far lower recombination rates, resulting in a clonal population structure and allowing characterization of the main phylogenetic groups (Stoppe et al., 2017). Several previous studies have reported that phylogenetic groups A and B1 are mostly commensal E. coli isolates. In this regard, Duriez et al. (2001) and Khairy et al. (2019) reported that phylogenetic groups A comprised the highest proportion of phylogenetic groups among human commensal E. coli and UPEC strains.
Certain papers have revealed that phylogroup A was the leading phylogroup in UPEC isolates (Grude et al., 2007; Romanus and Eze, 2011; Derakhshandeh et al., 2015; Khairy et al., 2019). Our study on the other hand implied that 18% of the isolates belonged to phylogenetic group A, which was greater than that in the studies performed in South Korea (3.44%) (Lee et al., 2016). Such observations indicated that we should consider all the potential risks of phylogenetic group A compared with those in phylogroup B2 (Marialouis and Santhanam, 2016). Moreover, the predominance of the phylogenetic group A in UPEC isolates, which is normally related to the commensal strains, implies that the gastrointestinal tract is the main origin of strains colonizing the urinary tracts (Moreno et al., 2006; Khairy et al., 2019).
Additionally, phylogroup D has been found to be the leading strain in certain studies (Themphachana et al., 2015; Gao et al., 2017); this reveals that the colon may be the main reservoir for strains causing UTI. Nevertheless, the second most prevalent group has been reported to be phylogenetic group D among drug-resistant UPEC strains, exhibiting slighter phylogenetic shift towards group B2 (Johnson et al., 2003; Adwan et al., 2015).
These variations in the occurrence of the phylogenetic groups may be on account of host genetic factors, site of infection, geographical distribution, or variations in methodology, the origin of isolates, and differences in the sample size. Additionally, these factors may be considered to be a source of heterogeneity. The present systematic review illustrated a significant heterogeneity among different phylogroups in the 28 studies conducted in Iran. We performed subgroup analysis of certain factors that confound the assessment (types of patients and geographical distribution based on region) and meta-regression for controlling this heterogeneity.
The geographical distribution of phylogenetic groups is variable in different regions of Iran. Accordingly, the subgroup analysis of the geographical distribution indicated that the predominant prevalence of phylogroups B2, D, A, and B1 had a frequency of 52%, 58%, 32%, and 10% in the north, southwest, northwest, and west of Iran, respectively.
Moreover, based on our findings, the high prevalence of phylogroup D was found among the isolates obtained from hospital-acquired infection rather than community-acquired infection. However, several reports have indicated a discrepancy in the prevalence of these phylogroups in both community and hospital infections. Meanwhile, our results revealed no significant differences on pooled prevalence of phylogroups neither in community-acquired nor hospital-acquired infections.
According to meta-regression, it seems as though the trend of phylogroup B2 incidence increased gradually from 20% in 2014 up to 83% in 2020. This increasing trend seems to be directly linked to the increased UPEC infections in phylogroup B2.
Investigations have shown that we have been experiencing an increase in the frequency of members of virulent phylogroups from clinical samples since 2009 to date.
Once rank correlation approaches show bias, there are possibly minor study effects. Meanwhile, according to the meta-regression analysis, the weight of the studies should not be regarded as a confounding factor. In addition, based on the sensitivity analysis, the exclusion of any works does not have any considerable impacts on the approximated pooled prevalence.
Furthermore, a superior characteristic is presented by these phylogenetic groups due to their partially higher content of virulence factors, which makes them virulent clinical isolates and harder to treat. In our study, the occurrence of VFs was greater in within-group B2 isolates taken from the patients’ urine compared with the other phylogenetic groups. This is consistent with former studies performed in South Korea (Lee et al., 2016), Denmark (Ejrnæs et al., 2011), Pakistan (Bashir et al., 2012), Ethiopia (Dadi et al., 2020), Mexico (Paniagua-Contreras et al., 2017), and Poland (Kot et al., 2016).
Based on the VF distribution in the phylogenetic groups, the existence of some genes had a high incidence in groups B2, D, B1, and A compared with that in the other VFs, including fimH papC and iucD. In this regard, Karami et al. (2017) reported that iutA and papC are encoded on mobile elements or pathogenicity islands in uropathogenic strains.
In this regard, some reports conducted in Iran revealed that the most frequent PAI marker belonged to PAIIV536. This PAI marker contains iron uptake system encoding genes and appears to be vital for successful colonization and wellness of UPEC strains throughout the urinary tract. However, previous studies have reported that there are a lot of differences concerning PAI markers among phylogenetic groups (Samei et al., 2016; Najafi et al., 2018). Accordingly, Najafi et al. (2018) reported that the majority of the isolates belonging to phylogenetic group B2 had all the investigated PAI markers. Moreover, their findings revealed that B1, A, and D groups of UPEC isolates had fewer PAI markers (Najafi et al., 2018).
Therefore, the performance of virulence genes in group B2 strains is mostly over the pathogenicity islands in the chromosome; however, virulence genes are often carried by group D strains, such as iutA, on plasmids. Nevertheless, group B2 is highly genetically diverse with at least nine subgroups (Le Gall et al., 2007), some of which may act well in the incorporation of genetic elements transferred horizontally compared with the others.
The differences concerning the host characteristics, geographical differences, and strain types are therefore responsible for variations in distributing such virulence factors in isolated UPEC. This result could be justified by the fact that the E. coli strain related to phylogroup B2 comprises a partially higher number of virulence genes compared with the E. coli related to the other phylogroups in other works on UPEC isolates.
In our work, group B2 isolates had high levels of resistance against ampicillin, cefepime, nalidixic acid, and ceftazidime and less resistance against nitrofurantoin and imipenem; meanwhile, a member of group D had a high resistance level against cefepime and ampicillin and less resistance against nitrofurantoin and amikacin. This finding is consistent with that of Bashir et al. (2012) and on the contrary to that of Iranpour et al. (2015) (Iran) who found a low drug resistance level for group D isolates.
Moreover, regarding antibiotic resistance, high levels of resistance was observed in our work among the members of phylogroups D, followed by B2, A, and B1.
Phylogenetic group D source was a considerable independent cause of antibiotic resistance, consistent with former studies, indicating that resistant genes could be achieved by isolates belonging to phylogenetic D.
There were certain limitations in our systematic review; primarily, phylogroups of UPEC have not yet been examined in numerous areas of Iran. Therefore, the frequency of phylogroups could not be completely represented. Furthermore, considering the heterogeneity found within the considered studies, the findings should be cautiously interpreted.
Conclusion
The results of the present study provided beneficial epidemiological information about the distribution of phylogroups in UPEC from Iranian patients. Our findings shed light on the fact that phylogroup B2 and group D were the most predominant phylogenetic groups among UPEC isolates in various regions of Iran, which is comparable with other parts of the world. Due to the relatively high frequency of phylogroup B2 and group D strains, it is necessary to pay attention to various groups involved in clinical care. Moreover, our results suggested that the members of phylogroup B2 strains may become reservoirs of genes encoding virulence factors. In addition, certain polygenetic groups were found to be more resistant than the others, which could be due to greater exposure of certain phylogenetic groups to antimicrobial agents. The dissemination of virulent phylogroups B2 and D could be suggested to be controlled through comprehensive infection control measures and through developing strategies for monitoring antibiotic therapy.
Author Contributions
Conceived and designed the experiments: RR and MH. Performed the experiments: MH, AF, and DZ Analyzed the data: AF and DZ. Contributed reagents/materials/analysis tools: MR and MH. Contributed to the writing of the manuscript: RR, AF, and MH. Manuscript revision and English editing: AP and MR. All authors read and approved the final manuscript.
Funding
This study was financially funded by the Babol University of Medical Sciences [Grant no. 724133882].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
Thanks for the guidance and advice from the “Clinical Research Development Unit of Baqiyatallah Hospital.”
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.790184/full#supplementary-material
References
- Adib N., Ghanbarpour R., Solatzadeh H., Alizade H. (2014). Antibiotic Resistance Profile and Virulence Genes of Uropathogenic Escherichia Coli Isolates in Relation to Phylogeny. Trop. Biomed. 31, 17–25. [PubMed] [Google Scholar]
- Adwan G., Issa B., Adwan K. (2015). Virulence Profile, Fluoroquinolone and Quinolone Resistance of Uropathogenic Escherichia Coli Isolates Recovered From Thabet Hospital-Tulkarm, Palestine. Microbiol. Res. J. Int. 5, 412–423. doi: 10.9734/BMRJ/2015/14293 [DOI] [Google Scholar]
- Alizade H., Ghanbarpour R., Aflatoonian M. R. (2014. a). Virulence Genotyping of Escherichia Coli Isolates From Diarrheic and Urinary Tract Infections in Relation to Phylogeny in Southeast of Iran. Trop. Biomed. 31, 174–182. [PubMed] [Google Scholar]
- Alizade H., Ghanbarpour R., Aflatoonian M. R., Abdollahi H. (2014. b). Determination of Phylogenetic Background, Fimbrial Genes, and Antibiotic Susceptibility of Escherichia Coli Isolates From Urinary Tract Infections in Bam Region, Iran. Comp. Clin. Pathol. 23, 1253–1257. doi: 10.1007/s00580-013-1771-z [DOI] [Google Scholar]
- Bahadori M., Motamedifar M., Derakhshandeh A., Firouzi R., Motamedi Boroojeni A., Alinejad M., et al. (2019). Genetic Relatedness of the Escherichia Coli Fecal Population and Strains Causing Urinary Tract Infection in the Same Host. MicrobiologyOpen 8, e00759. doi: 10.1002/mbo3.759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhtiari S., Mahmoudi H., Seftjani S. K., Amirzargar M. A., Ghiasvand S., Ghaffari M. E., et al. (2020). Antibiotic Resistance Pattern and Phylogenetic Groups of the Uropathogenic Escherichia Coli Isolates From Urinary Tract Infections in Hamedan, West of Iran. Iranian. J. Microbiol. 12, 388–394. doi: 10.18502/ijm.v12i5.4598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir S., Haque A., Sarwar Y., Ali A., Anwar M. I. (2012). Virulence Profile of Different Phylogenetic Groups of Locally Isolated Community Acquired Uropathogenic E. Coli From Faisalabad Region of Pakistan. Ann. Clin. Microbiol. Antimicrob. 11, 23. doi: 10.1186/1476-0711-11-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg C. B. (1985). A Measure to Aid in the Interpretation of Published Clinical Trials. Stat Med. 4, 1–9. doi: 10.1002/sim.4780040103 [DOI] [PubMed] [Google Scholar]
- Caméléna F., Birgy A., Smail Y., Courroux C., Mariani-Kurkdjian P., Le Hello S., et al. (2019). Rapid and Simple Universal Escherichia Coli Genotyping Method Based on Multiple-Locus Variable-Number Tandem-Repeat Analysis Using Single-Tube Multiplex PCR and Standard Gel Electrophoresis. Appl. Environ. Microbiol. 85, 2812–2818. doi: 10.1128/aem.02812-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clermont O., Christenson J. K., Denamur E., Gordon D. M. (2013). The Clermont Escherichia Coli Phylo-Typing Method Revisited: Improvement of Specificity and Detection of New Phylo-Groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019 [DOI] [PubMed] [Google Scholar]
- Cristea V. C., Gheorghe I., Czobor Barbu I., Popa L. I., Ispas B., Grigore G. A., et al. (2019). Snapshot of Phylogenetic Groups, Virulence, and Resistance Markers in Escherichia Coli Uropathogenic Strains Isolated From Outpatients With Urinary Tract Infections in Bucharest, Romania. BioMed. Res. Int. 2019, 5712371. doi: 10.1155/2019/5712371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadi B. R., Abebe T., Zhang L., Mihret A., Abebe W., Amogne W. (2020). Distribution of Virulence Genes and Phylogenetics of Uropathogenic Escherichia Coli Among Urinary Tract Infection Patients in Addis Ababa, Ethiopia. BMC Infect. Dis. 20, 108. doi: 10.1186/s12879-020-4844-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derakhshandeh A., Firouzi R., Motamedifar M., Arabshahi S., Novinrooz A., Boroojeni A. M., et al. (2015). Virulence Characteristics and Antibiotic Resistance Patterns Among Various Phylogenetic Groups of Uropathogenic Escherichia Coli Isolates. Jpn. J. Infect. Dis. 68, 428–431. doi: 10.7883/yoken.JJID.2014.327 [DOI] [PubMed] [Google Scholar]
- Dixit P. D., Pang T. Y., Studier F. W., Maslov S. (2015). Recombinant Transfer in the Basic Genome of Escherichia Coli. Proc. Natl. Acad. Sci. U. S. A. 112, 9070–9075. doi: 10.1073/pnas.1510839112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois D., Delmas J., Cady A., Robin F., Sivignon A., Oswald E., et al. (2010). Cyclomodulins in Urosepsis Strains of Escherichia Coli. J. Clin. Microbiol. 48, 2122–2129. doi: 10.1128/jcm.02365-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duriez P., Clermont O., Bonacorsi S., Bingen E., Chaventré A., Elion J., et al. (2001). Commensal Escherichia Coli Isolates are Phylogenetically Distributed Among Geographically Distinct Human Populations. Microbiol. (Reading England). 147, 1671–1676. doi: 10.1099/00221287-147-6-1671 [DOI] [PubMed] [Google Scholar]
- Ejrnæs K., Stegger M., Reisner A., Ferry S., Monsen T., Holm S. E., et al. (2011). Characteristics of Escherichia Coli Causing Persistence or Relapse of Urinary Tract Infections: Phylogenetic Groups, Virulence Factors and Biofilm Formation. Virulence 2, 528–537. doi: 10.4161/viru.2.6.18189 [DOI] [PubMed] [Google Scholar]
- Farajzadah Sheikh A., Goodarzi H., Yadyad M. J., Aslani S., Amin M., Jomehzadeh N., et al. (2019). Virulence-Associated Genes and Drug Susceptibility Patterns of Uropathogenic Escherichia Coli Isolated From Patients With Urinary Tract Infection. Infect. Drug Resist. 12, 2039–2047. doi: 10.2147/idr.s199764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayyazi A., Halaji M., Sadeghi A., Havaei S. A. (2020). High Frequency of Integrons and Efflux Pump in Uropathogenic Escherichia Coli Isolated From Iranian Kidney and non-Kidney Transplant Patients. Gene Rep. 21, 100873. doi: 10.1016/j.genrep.2020.100873 [DOI] [Google Scholar]
- Foxman B. (2014). Urinary Tract Infection Syndromes: Occurrence, Recurrence, Bacteriology, Risk Factors, and Disease Burden. Infect. Dis. Clinics North Am. 28, 1–13. doi: 10.1016/j.idc.2013.09.003 [DOI] [PubMed] [Google Scholar]
- Gao Q., Zhang D., Ye Z., Zhu X., Yang W., Dong L., et al. (2017). Virulence Traits and Pathogenicity of Uropathogenic Escherichia Coli Isolates With Common and Uncommon O Serotypes. Microbial. Pathogene. 104, 217–224. doi: 10.1016/j.micpath.2017.01.027 [DOI] [PubMed] [Google Scholar]
- Grude N., Potaturkina-Nesterova N. I., Jenkins A., Strand L., Nowrouzian F. L., Nyhus J., et al. (2007). A Comparison of Phylogenetic Group, Virulence Factors and Antibiotic Resistance in Russian and Norwegian Isolates of Escherichia Coli From Urinary Tract Infection. Clin. Microbiol. Infect. 13, 208–211. doi: 10.1111/j.1469-0691.2006.01584.x [DOI] [PubMed] [Google Scholar]
- Halaji M., Feizi A., Mirzaei A., Sedigh Ebrahim-Saraie H., Fayyazi A., Ashraf A., et al. (2020). The Global Prevalence of Class 1 Integron and Associated Antibiotic Resistance in Escherichia Coli From Patients With Urinary Tract Infections, a Systematic Review and Meta-Analysis. Microbial. Drug Resist. (Larchmont N.Y.). 26, 1208–1218. doi: 10.1089/mdr.2019.0467 [DOI] [PubMed] [Google Scholar]
- Hashemizadeh Z., Kalantar-Neyestanaki D., Mansouri S. (2017). Association Between Virulence Profile, Biofilm Formation and Phylogenetic Groups of Escherichia Coli Causing Urinary Tract Infection and the Commensal Gut Microbiota: A Comparative Analysis. Microbial. Pathogene. 110, 540–545. doi: 10.1016/j.micpath.2017.07.046 [DOI] [PubMed] [Google Scholar]
- Hemati Z., Ghanbarpour R., Alizade H. (2014). The Distribution of Beta Lactamase Genes in Escherichia Coli Phylotypes Isolated From Diarrhea and UTI Cases in Northwest Iran. Adv. Clin. Exp. Med. 23, 523–529. doi: 10.17219/acem/37217 [DOI] [PubMed] [Google Scholar]
- Hojabri Z., Mirmohammadkhani M., Kamali F., Ghassemi K., Taghavipour S., Pajand O. (2017). Molecular Epidemiology of Escherichia Coli Sequence Type 131 and its H30/H30-Rx Subclones Recovered From Extra-Intestinal Infections: First Report of OXA-48 Producing ST131 Clone From Iran. Eur. J. Clin. Microbiol. Infect. Dis. 36, 1859–1866. doi: 10.1007/s10096-017-3021-9 [DOI] [PubMed] [Google Scholar]
- Iranpour D., Hassanpour M., Ansari H., Tajbakhsh S., Khamisipour G., Najafi A. (2015). Phylogenetic Groups of Escherichia Coli Strains From Patients With Urinary Tract Infection in Iran Based on the New Clermont Phylotyping Method. BioMed. Res. Int. 2015, 846219. doi: 10.1155/2015/846219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. R., Delavari P., Kuskowski M., Stell A. L. (2001). Phylogenetic Distribution of Extraintestinal Virulence-Associated Traits in Escherichia Coli. J. Infect. Dis. 183, 78–88. doi: 10.1086/317656 [DOI] [PubMed] [Google Scholar]
- Johnson J. R., Kuskowski M. A., Owens K., Gajewski A., Winokur P. L. (2003). Phylogenetic Origin and Virulence Genotype in Relation to Resistance to Fluoroquinolones and/or Extended-Spectrum Cephalosporins and Cephamycins Among Escherichia Coli Isolates From Animals and Humans. J. Infect. Dis. 188, 759–768. doi: 10.1086/377455 [DOI] [PubMed] [Google Scholar]
- Karami N., Wold A. E., Adlerberth I. (2017). Antibiotic Resistance is Linked to Carriage of papC and iutA Virulence Genes and Phylogenetic Group D Background in Commensal and Uropathogenic Escherichia Coli From Infants and Young Children. Eur. J. Clin. Microbiol. Infect. Dis. 36, 721–729. doi: 10.1007/s10096-016-2854-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazemnia A., Ahmadi M., Dilmaghani M. (2014). Antibiotic Resistance Pattern of Different Escherichia Coli Phylogenetic Groups Isolated From Human Urinary Tract Infection and Avian Colibacillosis. Iranian. Biomed. J. 18, 219–224. doi: 10.6091/ibj.1394.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khairy R. M., Mohamed E. S., Abdel Ghany H. M., Abdelrahim S. S. (2019). Phylogenic Classification and Virulence Genes Profiles of Uropathogenic E. Coli and Diarrhegenic E. Coli Strains Isolated From Community Acquired Infections. PloS One 14, e0222441. doi: 10.1371/journal.pone.0222441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kot B., Wicha J., Grużewska A., Piechota M., Wolska K., Obrębska M. (2016). Virulence Factors, Biofilm-Forming Ability, and Antimicrobial Resistance of Urinary Escherichia Coli Strains Isolated From Hospitalized Patients. Turkish. J. Med. Sci. 46, 1908–1914. doi: 10.3906/sag-1508-105 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Subhadra B., Son Y. J., Kim D. H., Park H. S., Kim J. M., et al. (2016). Phylogenetic Group Distributions, Virulence Factors and Antimicrobial Resistance Properties of Uropathogenic Escherichia Coli Strains Isolated From Patients With Urinary Tract Infections in South Korea. Lett. Appl. Microbiol. 62, 84–90. doi: 10.1111/lam.12517 [DOI] [PubMed] [Google Scholar]
- Le Gall T., Clermont O., Gouriou S., Picard B., Nassif X., Denamur E., et al. (2007). Extraintestinal Virulence Is a Coincidental by-Product of Commensalism in B2 Phylogenetic Group Escherichia Coli Strains. Mol. Biol. Evol. 24, 2373–2384. doi: 10.1093/molbev/msm172 [DOI] [PubMed] [Google Scholar]
- Lewis A. J., Richards A. C., Mulvey M. A. (2016). Invasion of Host Cells and Tissues by Uropathogenic Bacteria. Microbiol. Spectr. 4, 359–381. doi: 10.1128/microbiolspec.UTI-0026-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marialouis X. A., Santhanam A. (2016). Antibiotic Resistance, RAPD- PCR Typing of Multiple Drug Resistant Strains of Escherichia Coli From Urinary Tract Infection (UTI). J. Clin. Diagn. Res. JCDR. 10, Dc05–Dc09. doi: 10.7860/jcdr/2016/16470.7389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moez N. M., Mashouf R. Y., Sedighi I., Shokoohizadeh L., Taheri M. (2020). Phylogroup Classification and Investigation the Relationships Between Phylogroups and Antibiotic Resistance Patterns of Uropathogenic E. Coli Isolated From Pediatric Urinary Tract Infection. Gene Rep. 20, 100758. doi: 10.1016/j.genrep.2020.100758 [DOI] [Google Scholar]
- Molina-López J., Aparicio-Ozores G., Ribas-Aparicio R. M., Gavilanes-Parra S., Chávez-Berrocal M. E., Hernández-Castro R., et al. (2011). Drug Resistance, Serotypes, and Phylogenetic Groups Among Uropathogenic Escherichia Coli Including O25-ST131 in Mexico City. J. Infect. Develop. Countries. 5, 840–849. doi: 10.3855/jidc.1703 [DOI] [PubMed] [Google Scholar]
- Momtaz H., Karimian A., Madani M., Safarpoor Dehkordi F., Ranjbar R., Sarshar M., et al. (2013). Uropathogenic Escherichia Coli in Iran: Serogroup Distributions, Virulence Factors and Antimicrobial Resistance Properties. Ann. Clin. Microbiol. Antimicrob. 12, 8. doi: 10.1186/1476-0711-12-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno E., Andreu A., Pérez T., Sabaté M., Johnson J. R., Prats G. (2006). Relationship Between Escherichia Coli Strains Causing Urinary Tract Infection in Women and the Dominant Faecal Flora of the Same Hosts. Epidemiol. Infect. 134, 1015–1023. doi: 10.1017/s0950268806005917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkhdelger Y., Gunregjav N., Dorjpurev A., Juniichiro N., Sarantuya J. (2017). Detection of Virulence Genes, Phylogenetic Group and Antibiotic Resistance of Uropathogenic Escherichia Coli in Mongolia. J. Infect. Develop. Countries. 11, 51–57. doi: 10.3855/jidc.7903 [DOI] [PubMed] [Google Scholar]
- Najafi A., Hasanpour M., Askary A., Aziemzadeh M., Hashemi N. (2018). Distribution of Pathogenicity Island Markers and Virulence Factors in New Phylogenetic Groups of Uropathogenic Escherichia Coli Isolates. Folia Microbiol. 63, 335–343. doi: 10.1007/s12223-017-0570-3 [DOI] [PubMed] [Google Scholar]
- Navidinia M., Najar Peerayeh S., Fallah F., Bakhshi B. (2013). Phylogenetic Groups and Pathogenicity Island Markers in Escherichia Coli Isolated From Children. Jundishapur. J. Microbiol. 6, e8362. doi: 10.5812/jjm.8362 [DOI] [Google Scholar]
- Navidinia M., Teymouri A. R., Goudarzi M. (2018). Assessment of Correlation Between Urinary Secretary IgA (Siga) Levels and Different Types of Urinary Tract Infection (UTI) in Various Age Groups. Arch. Adv. Biosci. 9, 45–49. doi: 10.22037/jps.v9i1.16481 [DOI] [Google Scholar]
- Norouzian H., Katouli M., Shahrokhi N., Sabeti S., Pooya M., Bouzari S. (2019). The Relationship Between Phylogenetic Groups and Antibiotic Susceptibility Patterns of Escherichia Coli Strains Isolated From Feces and Urine of Patients With Acute or Recurrent Urinary Tract Infection. Iranian. J. Microbiol. 11, 478–487. doi: 10.18502/ijm.v11i6.2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyaga V. N., Arbyn M., Aerts M. (2014). Metaprop: A Stata Command to Perform Meta-Analysis of Binomial Data. Arch. Public Health = Arch. Belges. Sante. Publique. 72, 39. doi: 10.1186/2049-3258-72-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua-Contreras G. L., Monroy-Pérez E., Rodríguez-Moctezuma J. R., Domínguez-Trejo P., Vaca-Paniagua F., Vaca S. (2017). Virulence Factors, Antibiotic Resistance Phenotypes and O-Serogroups of Escherichia Coli Strains Isolated From Community-Acquired Urinary Tract Infection Patients in Mexico. J. Microbiol. Immunol. Infect. = Wei. Mian. Yu. Gan. Ran. Za. Zhi. 50, 478–485. doi: 10.1016/j.jmii.2015.08.005 [DOI] [PubMed] [Google Scholar]
- Picard B., Garcia J. S., Gouriou S., Duriez P., Brahimi N., Bingen E., et al. (1999). The Link Between Phylogeny and Virulence in Escherichia Coli Extraintestinal Infection. Infect. Immun. 67, 546–553. doi: 10.1128/iai.67.2.546-553.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeispour M., Ranjbar R. (2018). Antibiotic Resistance, Virulence Factors and Genotyping of Uropathogenic Escherichia Coli Strains. Antimicrob. Resist. Infect. Control. 7, 118. doi: 10.1186/s13756-018-0411-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahdar M., Rashki A., Miri H. R., Ghalehnoo M. R. (2015). Detection of Pap, Sfa, Afa, Foc, and Fim Adhesin-Encoding Operons in Uropathogenic Escherichia Coli Isolates Collected From Patients With Urinary Tract Infection. Jundishapur. J. Microbiol. 8, e22647. doi: 10.5812/jjm.22647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos N. L., Dzung D. T. N., Stopsack K., Jankó V., Pourshafie M. R., Katouli M., et al. (2011). Characterisation of Uropathogenic Escherichia Coli From Children With Urinary Tract Infection in Different Countries. Eur. J. Clin. Microbiol. Infect. Dis. 30, 1587–1593. doi: 10.1007/s10096-011-1264-4 [DOI] [PubMed] [Google Scholar]
- Ranjbar R., Farahani O. (2018). The Prevalence of Virulence Genes and Virulotypes of Escherichia Coli Strains Isolated From Hospital Wastewaters in Tehran, Iran. Iran J. Public Health 47, 713–719. [PMC free article] [PubMed] [Google Scholar]
- Ranjbar R., Nazari S., Farahani O. (2020). Phylogenetic Analysis and Antimicrobial Resistance Profiles of Escherichia Coli Strains Isolated From Uti-Suspected Patients. Iranian. J. Public Health 49, 1743–1749. doi: 10.18502/ijph.v49i9.4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanus I. I., Eze A. T. (2011). Antibiotics Susceptibility Patterns and Clonal Relatedness of Uropathogenic Escherichia Coli in Abakaliki, Ebonyi State. Canad. J. Pure. Appl. Sci. 5, 1475–1479. [Google Scholar]
- Sadeghi A., Halaji M., Fayyazi A., Havaei S. A. (2020). Characterization of Plasmid-Mediated Quinolone Resistance and Serogroup Distributions of Uropathogenic Escherichia Coli Among Iranian Kidney Transplant Patients. BioMed. Res. Int. 2020, 2850183. doi: 10.1155/2020/2850183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehzadeh A., Zamani H. (2018). Characterization of (Uropathogenic) E. Coli Isolated From Urinary Tract Infections: Phylogenetic Typing and Distribution of Virulence-Associated Traits. Br. J. Biomed. Sci. 75, 40–42. doi: 10.1080/09674845.2017.1336834 [DOI] [PubMed] [Google Scholar]
- Salmani H., Azarnezhad A., Fayazi M. R., Hosseini A. (2016). Pathotypic and Phylogenetic Study of Diarrheagenic Escherichia Coli and Uropathogenic E. Coli Using Multiplex Polymerase Chain Reaction. Jundishapur. J. Microbiol. 9, e28331. doi: 10.5812/jjm.28331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samei A., Haghi F., Zeighami H. (2016). Distribution of Pathogenicity Island Markers in Commensal and Uropathogenic Escherichia Coli Isolates. Folia Microbiol. 61, 261–268. doi: 10.1007/s12223-015-0433-8 [DOI] [PubMed] [Google Scholar]
- Shahin N. P., Majid E., Amin T. B. A., Bita B. (2019). Host Characteristics and Virulence Typing of Escherichia Coli Isolated From Diabetic Patients. Gene Rep. 15, 100371. doi: 10.1016/j.genrep.2019.100371 [DOI] [Google Scholar]
- Smelov V., Naber K., Johansen T. E. B. (2016). Improved Classification of Urinary Tract Infection: Future Considerations. Eur. Urol. Supplements. 15, 71–80. doi: 10.1016/j.eursup.2016.04.002 [DOI] [Google Scholar]
- Sohrabi R., Zeighami H. (2016). Determination of Phylogenetic Groups and Antibiotic Resistance in Uropathogenic and Commensal Escherichia Coli Isolated From Patients in Zanjan City. J. Zanjan. Univ. Med. Sci. Health Services. 24, 107–118. [Google Scholar]
- Staji H., Rassouli M., Jourablou S. (2019). Comparative Virulotyping and Phylogenomics of Escherichia Coli Isolates From Urine Samples of Men and Women Suffering Urinary Tract Infections. Iranian. J. Basic. Med. Sci. 22, 211–214. doi: 10.22038/ijbms.2018.28360.6880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppe N., d C., Silva J. S., Carlos C., Sato M. I. Z., Saraiva A. M., et al. (2017). Worldwide Phylogenetic Group Patterns of Escherichia Coli From Commensal Human and Wastewater Treatment Plant Isolates. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabasi M., Asadi Karam M. R., Habibi M., Yekaninejad M. S., Bouzari S. (2015). Phenotypic Assays to Determine Virulence Factors of Uropathogenic Escherichia Coli (UPEC) Isolates and Their Correlation With Antibiotic Resistance Pattern. Osong. Public Health Res. Perspect. 6, 261–268. doi: 10.1016/j.phrp.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O., Skurnik D., Picard B., Denamur E. (2010). The Population Genetics of Commensal Escherichia Coli. Nat. Rev. Microbiol. 8, 207–217. doi: 10.1038/nrmicro2298 [DOI] [PubMed] [Google Scholar]
- Terlizzi M. E., Gribaudo G., Maffei M. E. (2017). UroPathogenic Escherichia Coli (UPEC) Infections: Virulence Factors, Bladder Responses, Antibiotic, and Non-Antibiotic Antimicrobial Strategies. Front. Microbiol. 8, 1566. doi: 10.3389/fmicb.2017.01566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Themphachana M., Kongpheng S., Rattanachuay P., Khianngam S., Singkhamanan K., Sukhumungoon P. (2015). Molecular Characterization of Virulence and Antimicrobial Susceptibility Profiles of Uropathogenic Escherichia Coli From Patients in a Tertiary Hospital, Southern Thailand. Southeast. Asian J. Trop. Med. Public Health 46, 1021. [PubMed] [Google Scholar]
- Touchon M., Perrin A., de Sousa J. A. M., Vangchhia B., Burn S., O’Brien C. L., et al. (2020). Phylogenetic Background and Habitat Drive the Genetic Diversification of Escherichia Coli. PloS Genet. 16, e1008866. doi: 10.1371/journal.pgen.1008866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazdanpour Z., Tadjrobehkar O., Shahkhah M. (2020). Significant Association Between Genes Encoding Virulence Factors With Antibiotic Resistance and Phylogenetic Groups in Community Acquired Uropathogenic Escherichia Coli Isolates. BMC Microbiol. 20, 241. doi: 10.1186/s12866-020-01933-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Zhang Y., Kwong J. S., Zhang C., Li S., Sun F., et al. (2015). The Methodological Quality Assessment Tools for Preclinical and Clinical Studies, Systematic Review and Meta-Analysis, and Clinical Practice Guideline: A Systematic Review. J. Evidence-Based Med. 8, 2–10. doi: 10.1111/jebm.12141 [DOI] [PubMed] [Google Scholar]
- Zhao R., Shi J., Shen Y., Li Y., Han Q., Zhang X., et al. (2015). Phylogenetic Distribution of Virulence Genes Among ESBL-Producing Uropathogenic Escherichia Coli Isolated From Long-Term Hospitalized Patients. J. Clin. Diagn. Res. JCDR. 9, Dc01–Dc04. doi: 10.7860/jcdr/2015/13234.6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.