Abstract
Pulmonary surfactant is a lipoprotein complex essential for lung function, and insufficiency or altered surfactant composition is associated with major lung diseases, such as acute respiratory distress syndromes, idiopathic pulmonary fibrosis, and chronic obstructive pulmonary disease. Pulmonary surfactant is primarily composed of phosphatidylcholine (PC) in complex with specialized surfactant proteins and secreted by alveolar type 2 (AT2) cells. Surfactant homeostasis on the alveolar surface is balanced by the rates of synthesis and secretion with reuptake and recycling by AT2 cells, with some degradation by pulmonary macrophages and loss up the bronchial tree. However, whether phospholipid (PL) transporters exist in AT2 cells to mediate reuptake of surfactant PL remains to be identified. Here, we demonstrate that major facilitator superfamily domain containing 2a (Mfsd2a), a sodium-dependent lysophosphatidylcholine (LPC) transporter, is expressed at the apical surface of AT2 cells. A mouse model with inducible AT2 cell–specific deficiency of Mfsd2a exhibited AT2 cell hypertrophy with reduced total surfactant PL levels because of reductions in the most abundant surfactants, PC containing dipalmitic acid, and PC species containing the omega-3 fatty acid docosahexaenoic acid. These changes in surfactant levels and composition were mirrored by similar changes in the AT2 cell lipidome. Mechanistically, direct tracheal instillation of fluorescent LPC and PC probes indicated that Mfsd2a mediates the uptake of LPC generated by pulmonary phospholipase activity in the alveolar space. These studies reveal that Mfsd2a-mediated LPC uptake is quantitatively important in maintaining surfactant homeostasis and identify this lipid transporter as a physiological component of surfactant recycling.
Keywords: omega-3 fatty acid, docosahexaenoic acid, phospholipid, blood–brain barrier, transporters
Abbreviations: AA, arachidonic acid; AT1, alveolar epithelial type 1; AT2, alveolar epithelial type 2; BAL, bronchioalveolar lavage; BSA, bovine serum albumin; DAG, diacylglycerol; DHA, docosahexaenoic acid; DPPC, dipalmitoylphosphatidylcholine; FA, fatty acid; IS, internal standard; LB, lamellar body; LPC, lysophosphatidylcholine; LPCAT1, lysophosphatidylcholine acyltransferase 1; Mfsd2a, major facilitator superfamily domain containing 2a; NBD, [12-(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]-dodecanoyl; PC, phosphatidylcholine; PFA, paraformaldehyde; PG, phosphatidylglycerol; PI, phosphatidylinositol; PL, phospholipid; PLA2, phospholipase A2; PUFA, polyunsaturated fatty acid; QC, quality control; RFP, red fluorescent protein; RT, room temperature; SP, surfactant protein; sPLA, secretory phospholipase; sPLA2, secretory PLA2; TBS, Tris-buffered saline; Tx, Triton-X
Pulmonary surfactant is critical for lung compliance during breathing, gas exchange, maintaining pulmonary barrier function, and keeping the lung alveolar epithelium dry. Pulmonary surfactant achieves these functions because of its ability to reduce alveolar surface tension and create an air–water barrier. Surfactant insufficiency and altered surfactant composition is associated with acute respiratory distress syndromes (1), idiopathic pulmonary fibrosis (2), and chronic obstructive pulmonary disease (3). Alveolar epithelial type 1 (AT1) and type 2 (AT2) cells are the major cell types forming alveolar structures. AT1 cells are extremely thin and flat that constitute approximately 95% of the alveolar surface and enable the efficient exchange of oxygen and carbon dioxide with the surrounding capillaries (4). AT2 cells are the secretory powerhouse that produce pulmonary surfactant. Pulmonary surfactant is a lipoprotein complex made up of 80 to 90% phospholipid (PL) and 5 to 10% surfactant proteins (SPs), SSP-A, SP-B, SP-C, and SP-D (5, 6). The major surfactant PL is dipalmitoylphosphatidylcholine (DPPC). DPPC, which is uniquely enriched in pulmonary surfactant, contains two palmitic acyl chains that impart the ability to be packed at a high density at the air–liquid interface, an important biophysical property for pulmonary function (5). PLs containing unsaturated fatty acyl chains are minor PL species of surfactant (7, 8).
Given the clinical importance of pulmonary surfactant, research into surfactant biosynthesis and metabolism has been an area of intense study. Surfactant phosphatidylcholine (PC) biosynthesis in AT2 cells occurs by de novo biosynthesis via the CDP–choline pathway. Fatty acyl chain remodeling of unsaturated fatty acids (FAs) through Land's cycle activity leads to the synthesis of DPPC, which is transported into lysosome-like organelles called lamellar bodies (LBs) by the action of the surfactant transporter ABCA3 (9, 10). Thus, DPPC is a unique lipid of LBs. LBs are secreted at the apical surface of AT2 cells by exocytosis as large aggregate membrane bilayers, which are rapidly adsorbed as a PL monolayer to form a pulmonary surfactant film. Through the processes of ventilation, the surfactant film becomes disorganized. Such spent surfactant is cleared via endocytosis by alveolar macrophages (11) or directly taken up by AT2 cells (12) with some loss up the bronchial tree (13). A portion of the spent surfactant PL can also be taken up intact by AT2 cells (14), where it is hydrolyzed by cytosolic phospholipase A2 (PLA2) and then reacylated by lysophosphatidylcholine (LPC) acyltransferase 1 (LPCAT1) (15, 16) and recycled back into LBs for resecretion (17, 18). This process of surfactant synthesis, turnover, and recycling must be balanced to maintain surfactant levels in the lung (15, 19, 20, 21). Surfactant PL has also been shown to be hydrolyzed by secretory phospholipase (sPLA) in the alveolar space to produce LPCs. Since LPCs are at very low levels in surfactant (7, 8) and have detergent-like properties detrimental to surfactant function, it is plausible that a transport system exists to recycle LPCs back to the AT2 cell.
Major facilitator superfamily domain containing 2a (Mfsd2a) is a sodium-dependent lysolipid transporter that is highly expressed in the endothelium of the blood–brain and blood–retinal barriers (22, 23). LPCs produced by the liver circulate in plasma bound to albumin (24). We have shown previously that Mfsd2a is the primary transporter by which the brain and eye acquires the essential FA docosahexaenoic acid (DHA) in the form of LPC–DHA. Mfsd2a deficiency models have significantly reduced DHA levels in brain and retina and present with severe microcephaly (22, 23, 25, 26, 27), similar to humans with loss-of-function mutations in Mfsd2a who develop severe microcephaly and intellectual disability (a.k.a. familial microcephaly 15, autosomal) (28, 29, 30, 31). Although Mfsd2a preferentially transports LPC containing polyunsaturated fatty acid (PUFA) moieties, it also transports saturated LPCs like LPC palmitate (22, 32). Importantly, Mfsd2a expression is not only confined to the brain or eye vasculature but also found to be expressed in other cell types and tissues, such as liver, intestine, and lymphocytes (33, 34).
Here, we present evidence that Mfsd2a is enriched at the apical surface of AT2 cells, where it is quantitatively important for the reuptake of LPCs that are produced through the hydrolysis of surfactant PC by sPLA A2 (sPLA2). Mfsd2a deficiency specifically in AT2 cells leads to AT2 cell hypertrophy with a deficiency in steady-state surfactant levels and altered surfactant lipid composition.
Results
Mfsd2a is expressed in lung AT2 cells
To determine lung cell types that express Mfsd2a, we utilized a Mfsd2a lineage tracing mouse line harboring a tamoxifen-inducible cre (ERT2cre) at exon 1 of the Mfsd2a gene and crossed to a red fluorescent protein (RFP) reporter line (designated as Mfsd2aERT2creTdTomato) that we previously demonstrated to faithfully report endogenous Mfsd2a expression at the blood–brain barrier (35). Treating Mfsd2aERT2creTdTomato mice with tamoxifen resulted in RFP expression exclusively in AT2 cells (Fig. 1). This is in agreement with Mfsd2a being highly expressed AT2 cells in human and mouse lung (Tabula Sapiens Consortium; (36, 37, 38, 39)). To specifically determine the function of Mfsd2a in AT2 cells in the lung, we generated an inducible AT2-specific Mfsd2a deletion model using Sftpc-CreERT2 (Mfsd2afl/fl Sftpc-CreERT2, henceforth designated AT2aKO). We chose to focus on the study of an AT2-specific model rather than whole-body Mfsd2a knockout mice because the latter model presents with multiple phenotypes that could confound our studies, such as increased plasma levels of LPCs, markedly small size, and increased basal metabolic rate, and altered T-cell immune function (33, 34). Immunohistochemical staining for endogenous Mfsd2a confirmed localization of Mfsd2a to AT2 cells of the lungs of 2afl/fl controls and indicated that Mfsd2a was successfully deleted from AT2 cells in AT2aKO mice as early as 10 days following tamoxifen administration (Fig. 2A).
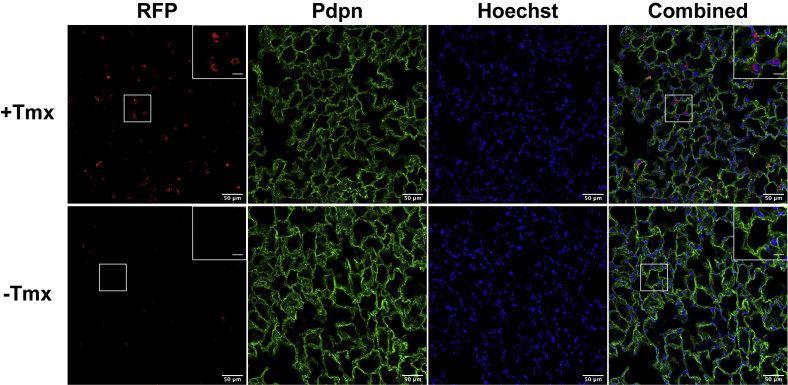
Figure 1.
Mfsd2aERT2creTdTomato reports Mfsd2a expression in lung AT2 cells. Immunohistochemical staining of RFP (red) on lung sections indicated Mfsd2a expression in AT2 cells with tamoxifen (+Tmx) treatment to induce RFP expression. Pdpn (green) stains AT1 cells. Lungs of untreated mice (−Tmx) served as a negative control. The scale bar represents 50 μm. Inset contains enlarged image indicated by white box. The scale bar represents 12.5 μm. Hoechst (blue) is a nuclear stain. Vehicle injected, n = 3; tamoxifen, n = 3. AT2, alveolar epithelial type 2; Mfsd2a, major facilitator superfamily domain containing 2a; RFP, red fluorescent protein.
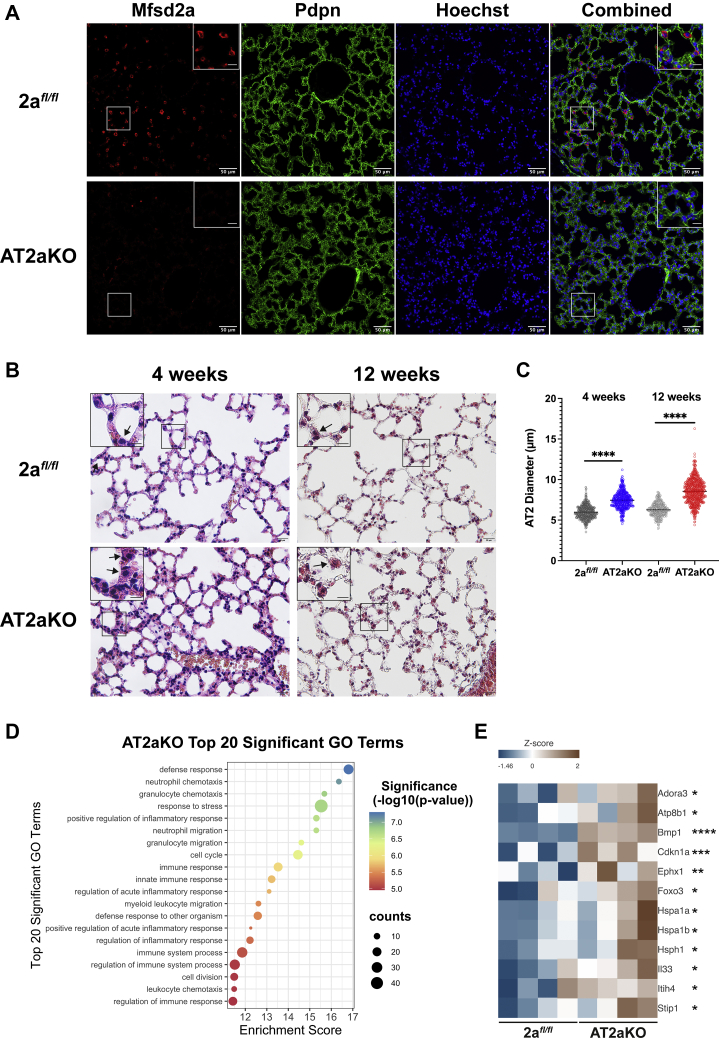
Figure 2.
Mfsd2a deficiency results in AT2 hypertrophy and changes to immune response and stress pathways.A, immunohistochemical staining on lung sections indicate Mfsd2a (red) expression in AT2 cells of 2afl/fl mice but absent in AT2aKO. Pdpn (green) stains AT1 cells. 2afl/fl, n = 6; AT2aKO, n = 4. The scale bar represents 50 μm. Inset in images is the enlarged area indicated by white box. The scale bar represents 12.5 μm. B, H&E-stained lung section of AT2aKO and 2afl/fl control 4 weeks post-tamoxifen. n = 6 per genotype. Masson's trichome-stained lung section of AT2aKO and 2afl/fl control 12 weeks post-tamoxifen. 2afl/fl, n = 3; AT2aKO, n = 7. AT2 cells are indicated by black arrows. The scale bar represents 20 μm. Inset in images is the enlarged area indicated by black box. The scale bar represents 10 μm. C, quantification of AT2 cell diameter from AT2aKO and 2afl/fl controls at 4 and 12 weeks post-tamoxifen represented as a scatterplot. Unpaired t test with Welch's correction, ∗∗∗∗p < 0.0001. Biological replicates as indicated in B. D, RNA-Seq analysis of AT2aKO and 2afl/fl AT2 cells. Bubble plot represents top 20 significantly upregulated or downregulated Gene Ontology terms. E, heatmap highlighting inflammatory and fibrosis markers that were upregulated in AT2aKO relative to 2afl/fl controls. Color bar indicates z-score transformation on median ratio normalized counts. n = 4 per genotype. Wald test, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. AT1, alveolar epithelial type 1; AT2, alveolar epithelial type 2; Mfsd2a, major facilitator superfamily domain containing 2a.
We next examined AT2aKO lungs for potential phenotypes as a consequence of Mfsd2a deficiency. Histological analysis at 4 weeks post-tamoxifen treatment showed no obvious structural changes or fibrosis in the lungs of AT2aKO mice relative to lungs of 2afl/fl control mice (Fig. 2B, left panel). However, AT2 cells of AT2aKO appeared hypertrophic relative to 2afl/fl controls (Fig. 2B, left panel, and Fig. 2C) and remained hypertrophic up to 3 months after deletion of Mfsd2a (Fig. 2B, right panel, and Fig. 2C). To further understand how Mfsd2a deficiency affects the lung and reveal potential adaptations to Mfsd2a deficiency, transcriptomic analysis by bulk RNA-Seq was carried out on AT2 cells isolated from AT2aKO and 2afl/fl controls. We identified 112 upregulated and 191 downregulated differentially expressed genes, overall indicating minor transcriptional changes in AT2 cells in response to Mfsd2a deficiency. Pathway analysis of differentially expressed genes between AT2aKO and 2afl/fl controls identified changes to immune response, response to stress, and cell cycle (Fig. 2D) with increased expression of inflammatory/chronic obstructive pulmonary disease markers in AT2aKO AT2 (Fig. 2E). Notably, Kennedy and CDP–diacylglycerol (DAG) pathway genes (Chka/b, Pcyt1a/b, Chpt1, Etnk1/2, Pcyt2, Ept1, Cds1/2, and Cdipt) and ABCA3 were not significantly changed in AT2aKO AT2 relative to 2afl/fl controls (see DESeq2 median ratio normalized counts available on GSE186170).
Mfsd2a transports LPC at the apical surface of AT2 cells
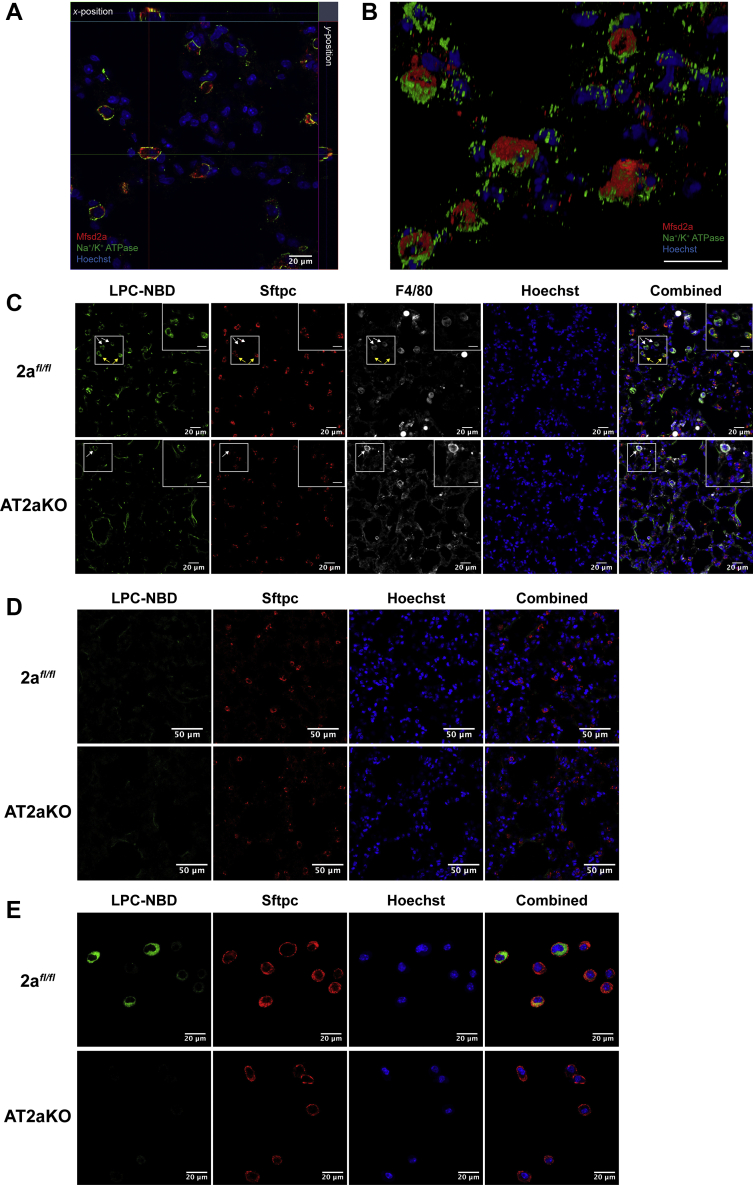
AT2 cells exhibit cell polarity with an apical membrane having microvilli facing the alveoli space and a basolateral membrane associated with AT1 cells facing the interstitial region. We next sought to determine if Mfsd2a expression is polarized in AT2 cells by generating 3D reconstructions of confocal images of lung sections stained for Mfsd2a and the AT2 basolateral membrane marker Na+/K+ ATPase α1. This image analysis indicated that Mfsd2a is enriched on the apical surface of AT2 cells (Fig. 3, A and B and Movie S1) and raises the hypothesis that Mfsd2a transports LPCs from the alveolar surface rather than from blood.
Figure 3.
Mfsd2a mediates the uptake of LPCs into AT2 at the apical surface.A, orthoview of z-stack of lung sections stained for Mfsd2a (red) and the AT2 basolateral membrane marker Na+/K+ ATPase α1 (green). The scale bar represents 20 μm. B, 3D reconstruction of confocal images in (A) indicated enrichment of Mfsd2a (red) at the apical side of AT2 cells. 2afl/fl, n = 6; AT2aKO, n = 4. The scale bar represents 50 μm. C, intratracheal instillation of LPC-NBD. Immunohistochemical staining of Sftpc (red) and F4/80 (gray) on lung sections indicates accumulation of LPC-NBD (green) only in 2afl/fl AT2 cells but absent in AT2aKO AT2. LPC-NBD accumulation in macrophages is observed in both AT2aKO and 2afl/fl AT2. The scale bar represents 20 μm. Inset in images is the enlarged area indicated by white box. The scale bar represents 10 μm. n = 4 per genotype. D, intravenous instillation of LPC-NBD. No LPC-NBD uptake was observed in AT2aKO or 2afl/fl AT2 cells. Sftpc (red) stains AT2 cells. n = 4 per genotype. The scale bar represents 50 μm. E, LPC-NBD (green) was taken up by AT2 cells isolated from 2afl/fl but not AT2aKO mice in vitro. Sftpc (red) denotes AT2 cells. Hoechst (blue) denotes nuclei. n = 5 per genotype. The scale bar represents 20 μm. AT2, alveolar epithelial type 2; LPC, lysophosphatidylcholine; Mfsd2a, major facilitator superfamily domain containing 2a; NBD, [12-(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]-dodecanoyl.
Given that Mfsd2a is enriched at the AT2 apical membrane, we sought to determine if it transports LPC from the alveolar surface. To do this, we utilized a fluorescently labeled LPC with a 16:1 fatty acyl chain (LPC-[12-(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]-dodecanoyl [NBD]) that is transported by Mfsd2a (40). LPC-NBD was instilled directly into the trachea of AT2aKO and 2afl/fl controls. Histological analysis indicated that LPC-NBD accumulated in both AT2 and macrophages in 2afl/fl controls but only in macrophages of AT2aKO (Fig. 3C). These data indicate that Mfsd2a is functional at the apical side of AT2 cells to take up LPCs from the alveoli surface, whereas LPCs can be taken up by alveolar macrophages independent of Mfsd2a transport, possibly through endocytosis together with intact surfactant (14). We consistently found that the signal for Sftpc, an SP we used to identify AT2 cells, was found to significantly reduced in AT2aKO relative to 2afl/fl controls (Fig. S1, A and B). Moreover, Sftpc mRNA transcripts were reduced by 56% in AT2aKO (Fig. S1C). We suspected that the reduced levels of Sftpc might be due to the knockin of Cre-ERT2 at the Sftpc locus (41) and quantified Sftpc signal in AT2 cells of SftpcCre-ERT2 mice. Indeed, the levels of Sftpc signal were similarly reduced in this cre line relative to 2afl/fl controls, supporting the conclusion that reduced levels of Sftpc was due to transgene construction and unrelated to Mfsd2a deletion. It is also important to note that Sftpc deficiency is not essential for surfactant production (42). In contrast, the fluorescent signal of Sftpb, an SP essential for surfactant production and proteolytic processing of Sftbc (20), was similar in AT2 cells of 2afl/fl controls (Fig. S2, A and B), in agreement with similar levels of Sftpb in bronchial lavages from AT2aKO and 2afl/fl controls (Fig. S2C).
We next tested whether LPC uptake into lung from blood is mediated by Mfsd2a. Despite delivering more than three times the amount of LPC-NBD instilled intratracheally, LPC-NBD was not taken up by AT2 when delivered intravenously (Fig. 3D), consistent with the absence of Mfsd2a at the basolateral membrane of AT2 cells. To confirm that these findings are directly the consequence of Mfsd2a transport activity in AT2 cells, we isolated AT2 cells from the lungs of AT2aKO and 2afl/fl controls and tested for transport of LPC-NBD in vitro. Detection of Sftpc by immunofluorescence clearly indicated the identity of AT2 cells in vitro. Indeed, AT2 cells freshly isolated from 2afl/fl lungs, but not from AT2aKO lungs, exhibited uptake of LPC-NBD (Fig. 3E), demonstrating that the uptake of LPCs by AT2 cells is dependent on Mfsd2a.
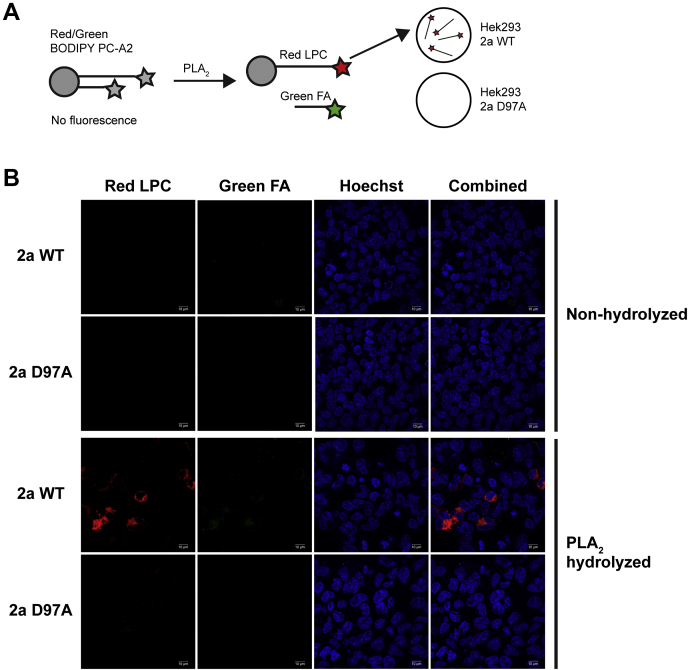
The uptake of LPC-NBD by Mfsd2a expressed on the apical surface of AT2 cells suggests that the origin of LPC in the alveolar space is through the hydrolysis of surfactant PC by sPLA2 produced by AT2 cells or other cell types such as macrophages (43). Given that sPLA2 activity has been demonstrated to contribute to surfactant PC turnover (43, 44, 45, 46), we set up experiments to test if LPC generated from sPLA2 hydrolysis of PC in vivo would be taken up by AT2 cells in an Mfsd2a-dependent fashion. To approach this experimentally, we made use of a commercially available Red/Green BODIPY PC-A2 probe that has a hydrolyzable ester bond only at the sn-2 position to release a red fluorescent BODIPY-labeled LPC and a green fluorescent BODIPY linked to a short-chain FA (Fig. 4A). Prior to hydrolysis by sPLA2, the dual Red/Green BODIPY is not fluorescent because of quenching due to the proximity of both red and green BODIPY moieties, thus allowing in vivo monitoring of sPLA2 activity. To determine if Mfsd2a is able to transport the red fluorescent LPC product derived from sPLA2 hydrolysis, we made use of cells overexpressing WT Mfsd2a or its corresponding transport-deficient mutant D97A (22, 32, 40). Red fluorescent LPC was taken up only in cells expressing WT Mfsd2a but not D97A (Fig. 4B). Moreover, the green fluorescent short-chain FA product was not taken up by cells (Fig. 4B). These data indicate that Red/Green BODIPY PC-A2 probe is a suitable fluorescent probe for quantifying LPC uptake by Mfsd2a that is generated by sPLA2 activity in vivo.
Figure 4.
Red fluorescent LPC product generated from PLA2action on Red/Green BODIPY PC-A2 is transported by Mfsd2a.A, model of experimental setup showing that Red/Green BODIPY PC-A2 probe is hydrolyzed by PLA2 to release a red fluorescent BODIPY-labeled LPC (red LPC) and a green fluorescent BODIPY linked to a short-chain FA (green FA). B, nonhydrolyzed or PLA2 hydrolyzed Red/Green BODIPY were added to human embryonic kidney 293 cells overexpressing WT Mfsd2a (2a WT) or transport-deficient mutant D97A (2a D97A). Red fluorescent LPC is taken up only in cells expressing WT Mfsd2a but not D97A. The green fluorescent short-chain FA product was not taken up by cells. This experiment was repeated twice with similar results. The scale bar represents 10 μm. FA, fatty acid; LPC, lysophosphatidylcholine; Mfsd2a, major facilitator superfamily domain containing 2a; PLA2, phospholipase A2.
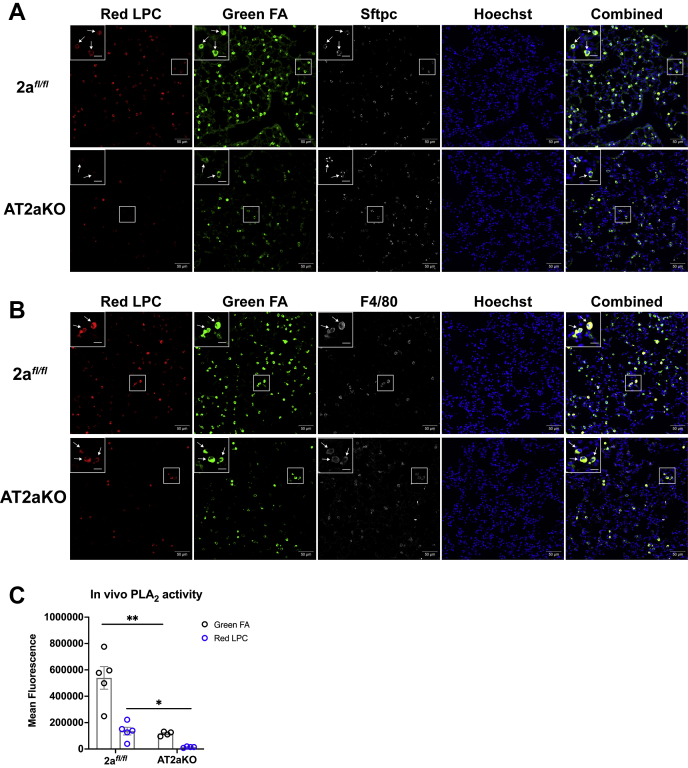
To assess sPLA2 activity in vivo, Red/Green BODIPY PC-A2 was instilled intratracheally into both AT2aKO and 2afl/fl controls, and 2 h later, lungs were harvested for processing and fluorescent imaging. In sections from 2afl/fl controls, both red and green fluorescence was seen in AT2 cells, whereas only green fluorescence was observed in AT2 cells of AT2aKO lungs (Fig. 5A). It is important to note that although red fluorescent LPC was taken up by cells in AT2aKO lungs, immunohistochemical staining with F4/80 revealed that these are macrophages and not AT2 cells, which were also observed in 2afl/fl macrophages (Fig. 5B). It is also notable that the overall fluorescence intensity of AT2aKO samples was significantly less than 2afl/fl controls (Fig. 5C), indicating that AT2aKO samples have reduced in vivo sPLA2 activity in the alveolar space.
Figure 5.
Uptake of LPC by AT2 cells is dependent on lung phospholipase A2(PLA2) activity.A, intratracheal instillation of Red/Green BODIPY PC-A2. Both red LPC and green FA are taken up by 2afl/fl AT2 as seen by colocalization with Sftpc (indicated by white arrows in insets of area denoted by white squares), whereas only green FA is taken up by AT2aKO AT2. B, red LPC is also taken up by macrophages as seen by colocalization with F4/80 (indicated by white arrows in insets of area denoted by white squares) in both AT2aKO and 2afl/fl AT2. C, quantification of red and green fluorescence in lung sections. 2afl/fl, n = 5; AT2aKO, n = 4. The scale bar represents 50 μm; the scale bar (inset) represents 12.5 μm. Unpaired t test with Welch's correction, ∗p < 0.05; ∗∗p < 0.01. AT2, alveolar epithelial type 2; FA, fatty acid; LPC, lysophosphatidylcholine.
We next asked if this reduced in vivo sPLA2 activity was due to reduced amounts of sPLA2 in bronchioalveolar lavage (BAL) of AT2aKO. To address this question, total sPLA2 activity was measured ex vivo on BAL isolated from AT2aKO and 2afl/fl controls using the Red/Green BODIPY PC-A2 probe, and lipids were extracted and resolved by thin layer chromatography (Fig. 6, A and B). In addition, we made use of a second commercially available Bis-BODIPY FL C11-PC probe that has hydrolyzable ester bonds at both the sn-1 and sn-2 positions to release a green fluorescent BODIPY-labeled LPC and a green fluorescent BODIPY linked to 11-carbon FA. Prior to hydrolysis by sPLA2, this probe is not fluorescent because of quenching due to the proximity of both BODIPY moieties. The nonhydrolyzed Red/Green BODIPY PC-A2 and Bis-BODIPY FL C11-PC probes (black arrows in Fig. 6, B and C, respectively) is fluorescent in thin layer chromatography analysis because the fluorescent moieties on each acyl chain are not ordered and quenched in organic solvents as they are in aqueous solutions like BAL in vivo. PLA2 activity was comparable between both genotypes regardless of the fluorescent probe used (Fig. 6D). Collectively, these data support a pathway by which LPCs generated by sPLA2 action on surfactant PC is taken up via Mfsd2a expressed on the apical surface of AT2 cells.
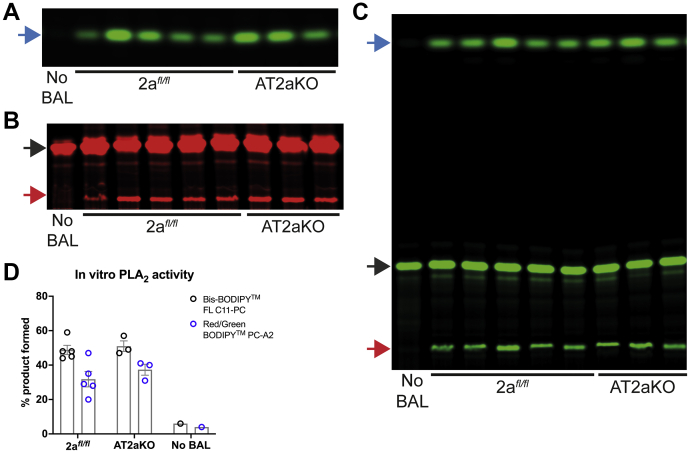
Figure 6.
Assessment of sPLA2activity in BAL. sPLA2 activity was measured in BAL isolated from AT2aKO or 2afl/fl using Red/Green BODIPY PC (A and B) or Bis-BODIPY FL C11-PC (C), and lipids were extracted and resolved by TLC. sPLA2 activity was comparable between both genotypes. LPC and FA were indicated by red and blue arrows, respectively. Unhydrolyzed PC substrates (no BAL, black arrows) are fluorescent in organic solvent used for TLC analysis as these substrates are no longer ordered and quenched as they are in BAL in vivo. 2afl/fl, n = 5; AT2aKO, n = 3. D, quantification of sPLA2 activity is represented as the mean ± SE of the ratio of the sum of products (LPC and FA) over the sum of products and unhydrolyzed substrate. 2afl/fl, n = 5; AT2aKO, n = 3. BAL, bronchioalveolar lavage; FA, fatty acid; FL, full-length; LPC, lysophosphatidylcholine; PC, phosphatidylcholine; sPLA2, secretory PLA2.
Mfsd2a deficiency reduces surfactant levels and alters surfactant composition
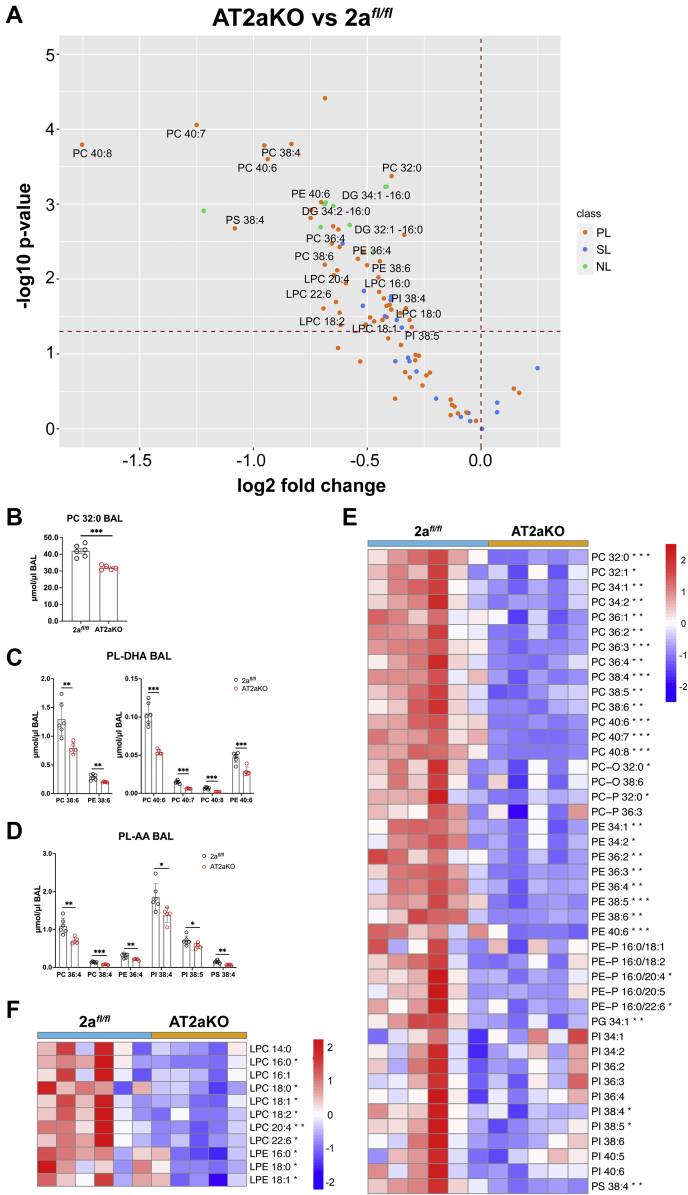
To determine if Mfsd2a deficiency affects surfactant levels and lipid composition, lipidomics was performed on BALs harvested from AT2aKO and 2afl/fl controls at 4 weeks post-tamoxifen treatment. We quantified 96 lipid species common to BAL (Table S1). Remarkably, major surfactant PL species were found to be reduced in AT2aKO BAL relative to 2afl/fl controls (Fig. 7A) including the major surfactant PC-32:0 (DPPC) (Fig. 7, A and B), which was reduced by 24% in AT2aKO BAL. Moreover, major PL species having the polyunsaturated fatty acyl chain DHA (PC–DHA) (Fig. 7, A and C) and arachidonic acid (PL-AA) (Fig. 7, A and D) were reduced by 38% and 29%, respectively, in AT2aKO BAL relative to 2afl/fl controls at 4 weeks post-tamoxifen treatment. Several minor PL species found in BAL were also significantly reduced in AT2aKO BAL (Fig. 7E). Supportive of reduced sPLA2 activity in vivo (Fig. 5), lyso-PL species were reduced in AT2aKO BAL relative to 2afl/fl controls (Fig. 7, A and F).
Figure 7.
Mfsd2a deficiency leads to reductions in total BAL phospholipids (PLs). Lipidomic analysis of AT2aKO and 2afl/fl BAL 4 weeks post-tamoxifen treatment. A, BAL lipid species represented as a volcano plot. Dots above dashed horizontal line indicate lipid species that are significantly different between AT2aKO and 2afl/fl AT2. Most of the lyso-PL and PL species are reduced in AT2aKO BAL relative to 2afl/fl control (PL, SL, and NL). Total PC 32:0 (B) and PL species containing DHA (PL-DHA) (C) or AA (PL-AA) (D) were reduced in AT2aKO BAL relative to 2afl/fl. BAL PLs (E) and lyso-PLs (F) represented as a heatmap. Color bar indicates z-score transformation on μmol/μl BAL PLs. Data for B–D are represented as mean μmol/μl BAL ± SE, with individual points denoting biological replicates. Fatty acid identity is designated as the number of carbons:number of double bonds. For all panels in this figure, 2afl/fl, n = 6; AT2aKO, n = 5. Unpaired t test with Welch's correction, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. AA, arachidonic acid; AT2, alveolar epithelial type 2; BAL, bronchioalveolar lavage; DHA, docosahexaenoic acid; Mfsd2a, major facilitator superfamily domain containing 2a; NL, neutral lipid; PC, phosphatidylcholine; SL, sphingolipid.
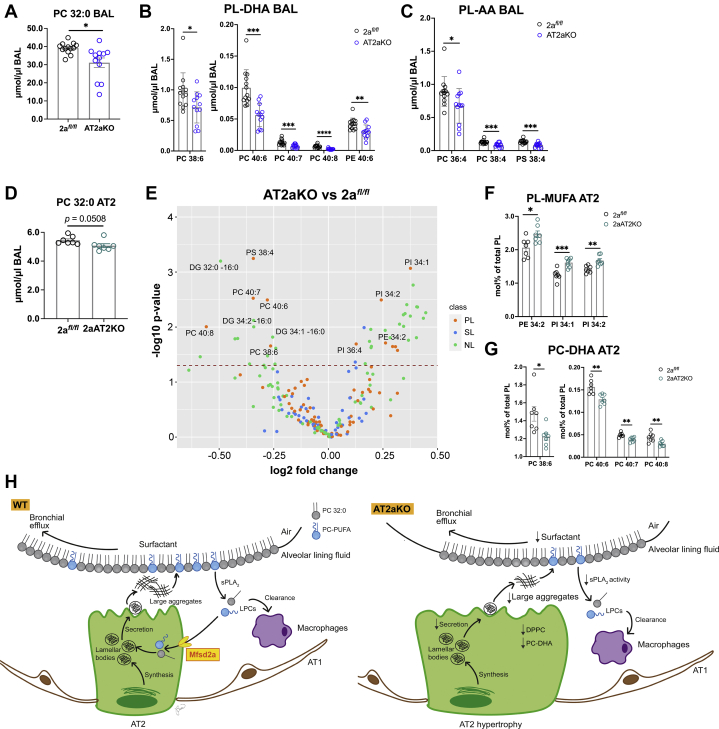
To know whether these PL changes in surfactant occur early after Mfsd2a deletion, lipidomics was performed on BAL harvested from lungs 2 weeks following tamoxifen induction (Table S2). Indeed, reductions in total DPPC (Fig. 8A), PL-DHA (Fig. 8B), and PL-AA (Fig. 8C) were significantly reduced in AT2aKO BAL relative to 2afl/fl controls. Quantification of 212 lipid species of the lipidome from isolated AT2 cells at this 2 week time point showed a reduction in total DPPC in AT2aKO relative to 2afl/fl controls (Fig. 8D and Table S3). This reduction in DPPC is particularly informative because DPPC is unique to surfactant found in LBs (47, 48) and consistent with reduced DPPC in BAL (Figs. 7B and 8A). AT2 cells from AT2aKO also exhibited an increase in mol% of PLs containing monounsaturated FAs (Fig. 7, E and F) and reductions in PC–DHA (Fig. 8, E and G) and phosphatidylserine 38:4 (Fig. 8E), revealing a shift toward more saturated PLs.
Figure 8.
Mfsd2a deficiency alters the AT2 cell lipidome reflective of changes in BAL. Lipidomic analysis of AT2aKO and 2afl/fl BAL 2 weeks post-tamoxifen treatment. Total PC 32:0 (A) and phospholipid (PL) species containing DHA (PL-DHA) (B) or AA (PL-AA) (C) were reduced in AT2aKO BAL relative to 2afl/fl. D, lipidomic analysis of AT2aKO and 2afl/fl AT2 cells 2 weeks post-tamoxifen treatment. Total PC 32:0 was reduced in AT2aKO AT2 cells relative to 2afl/fl. E, mol% lipid species represented as a volcano plot. Dots above dashed line indicate lipid species that are significantly different between AT2aKO and 2afl/fl AT2 (PL, SL, and NL. F, PLs with monounsaturated fatty acids (PL-MUFA) species were increased in AT2aKO AT2 cells relative to 2afl/fl. G, PC–DHA species were reduced in AT2aKO AT2 cells relative to 2afl/fl, similar to changes in the BAL lipidome. For A–D, data are represented as mean μmol/μl BAL ± SE, with individual points denoting biological replicates. 2afl/fl, n = 13; AT2aKO, n = 12. For F–H, data represented as mol% ± SE of PLs, with individual points denoting biological replicates. n = 7 per genotype. Unpaired t test with Welch's correction, ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. H, model to show Mfsd2a is required for surfactant homeostasis. AT2 cells synthesize surfactants, which are secreted as DPPC-rich lamellar bodies at the apical surface that unravel into large surfactant aggregates that make up the alveolar lining fluid. Spent surfactant is released as small surfactant aggregates that are hydrolyzed by sPLA2 in the alveolar space to produce lysophosphatidylcholines (LPCs). LPC are then taken up by Mfsd2a expressed at the apical surface of AT2 cells and serve as a precursor for the synthesis of PC–DHA that are then remodeled into the main surfactant PL DPPC. Some LPCs can also be cleared by alveolar macrophages. In the absence of Mfsd2a, uptake of LPCs is limited resulting in reduction in PC–DHA leading to reduced resynthesis of DPPC. Moreover, reduced sPLA2 activity in the absence of Mfsd2a might be a result of altered surfactant composition.
Discussion
LBs containing surfactant are secreted at the apical surface of AT2 cells, which unravel into large surfactant aggregates that partition into the surfactant reservoir. As the lung goes through repeated cycles of expansion and compression, hydrophobic SPs like SP-B and SP-C facilitate the insertion and spreading of PLs from the surfactant reservoir at the interface, whereas spent surfactant is released as small surfactant aggregates. Spent surfactant can either be hydrolyzed by secretory PLA2 (sPLA2) in the alveolar space to produce LPCs taken up intact by AT2 (14) and recycled back to LBs for resecretion or lost up the bronchial tree (13). Although it has been proposed that LPCs can be cleared by alveolar macrophages (11), it is possible that a transport system exists to recycle LPCs back to AT2 for the resynthesis of PCs. Here, we demonstrated that Mfsd2a, a sodium-dependent LPC transporter, is expressed at the apical surface of AT2 cells, where it plays a role in the uptake of LPCs produced at the alveolar surface and that this transport pathway is quantitatively important to maintain normal surfactant levels and composition.
AT2 cells have the machinery to de novo synthesize PC via the CDP–DAG pathway and are able to obtain free FAs directly from the plasma to be utilized for surfactant production (49). Lungs of mice deficient in the second enzymatic step in the CDP–DAG pathway (i.e., PCYT1A) developed normally but rapidly developed respiratory failure at birth because of surfactant deficiency (21), highlighting the importance of this de novo PC synthesis pathway. However, the rate of de novo surfactant synthesis is slow, supporting the requirement for recycling of spent surfactant to efficiently maintain surfactant homeostasis in the lung (44). The use of instillation of radiolabeled isotopic-labeled DPPC has indicated the existence of two pathways for the recycling of surfactant PC, with one being uptake of intact surfactant by AT2 cells through an unknown mechanism (14, 50) and another pathway leading to the formation of LPC through the action of sPLA2 (44). Our studies here support the physiological importance of the latter pathway indicating that uptake of LPC is mediated by Mfsd2a.
Lipidomic analysis on BAL and AT2 obtained from AT2aKO mice showed that not only was DPPC levels reduced but also total PL-DHA and PL-AA were also reduced in AT2aKO relative to 2afl/fl controls. Reductions in DPPC, which is the most abundant BAL PL, is intriguing. Given that intratracheal instillation of exogenous surfactant in lungs of mice did not reduce endogenous surfactant production (44) suggests a direct role of LPC uptake by Mfsd2a to be reacylated into PC to maintain surfactant levels in the lung. Moreover, reductions in DPPC, a specific lipid of the LB, were similarly observed in AT2aKO AT2 cells, suggesting reduced recycling of surfactant. That LPCAT1 knockout mice have reduced DPPC in BALF clearly supports the importance of PC remodeling in surfactant production and suggests that LPC uptake by Mfsd2a in AT2 cells likely is acylated by LPCAT1, the primary LPCAT expressed in lungs (15, 16). Moreover, the decrease in PC–PUFA in AT2aKO AT2 cells is reminiscent of DHA deficiency seen in brain and eye of various Mfsd2a knockout mouse models (22, 23, 25, 26, 27). Although PUFAs only constitute a small percentage of surfactant PC, they might be required for the remodeling of PC in AT2 cells (44) where LPC palmitate taken up by AT2 cells or DPPC endogenously synthesized by AT2 cells is remodeled by the action of PLA2 and LPCAT1, leading to the formation of fully saturated DPPC that is packaged into LBs and secreted as surfactant (10, 16, 18, 43).
How might Mfsd2a transport function in AT2 cells impact innate immunity in the lung? A closer examination of our lipidomic data revealed that surfactant of AT2aKO had significant reductions in phosphatidylglycerol (PG 34:1) and several species of phosphatidylinositol (PI 38:4 and PI 38:5; Fig. 7). These two anionic PLs are minor constituents of pulmonary surfactant, yet play important roles in innate immunity in the lung. Voelker et al. have demonstrated that PG (specifically 1-palmitoyl-2-oleoyl-sn-glycero-3-[phospho-rac-(1-glycerol)]) and PI antagonize toll-like receptor 4 and toll-like receptor 2 signaling in alveolar macrophages, respectively, to limit lung pathogenesis caused by respiratory syncytial virus and influenza A virus and raises the possibility that these PLs can have therapeutic utility (51, 52, 53, 54, 55, 56). Whether Mfsd2a deficiency will exacerbate pathology resulting from respiratory syncytial virus and influenza A virus remains to be determined.
Interestingly, sPLA2 activity appeared to be reduced in the alveolar space in AT2aKO lungs, whereas total sPLA2 activity in isolated BAL was similar in AT2aKO and 2afl/fl controls. The specific sPLA2 that generates LPC in the alveolar space is not known, but several sPLA2 enzymes have been shown to be expressed in the lung, namely PLA2G1B, PLA2G5, and PLA2G10 (57, 58, 59). It is known that sPLA2 is an interfacial enzyme whose kinetics can be effected by PL acyl chain composition and whether the PL substrate is in the form of large or small PL aggregates or as surfactant monolayers (60, 61, 62). The isolation of BAL for measuring total sPLA2 activity is expected to mix all surfactant contents and not reflect the actual in vivo physical state of surfactant. Therefore, we might predict that the reductions in DPPC and PL-PUFAs in BAL of AT2aKO might be causative for reduced sPLA2 activity that we observed in vivo (Fig. 5). Reduced sPLA2 activity might also be considered a positive adaptation to Mfsd2a deficiency and could explain why LPC levels were not increased in BAL in the absence of Mfsd2a. In summary, our findings support a model in which LPCs generated by sPLA2 activity are recycled by Mfsd2a in AT2 cells, a process that is physiologically important for surfactant homeostasis (Fig. 8H).
Experimental procedures
Mouse models
Experimental protocols involving mice were approved by SingHealth Institutional Animal Care and Use Committee. Adult mice were anesthetized with a combination of ketamine (20 mg/kg body weight) and xylazine (2 mg/kg body weight) for all experiments. Mfsd2a lineage tracing mouse line (designated as Mfsd2aERT2cre) was generated by genetically knocking in tamoxifen-inducible cre (ERT2cre) into exon 1 of the Mfsd2a gene. The Mfsd2aERT2cre line was then crossed to a cre recombinase–inducible TdTomato reporter line (B6;129S6-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J; The Jackson Laboratory) (63). Mfsd2a floxed mice (2afl/fl) were generated as described previously (23). Tamoxifen-inducible AT2-specific knockout mice (AT2aKO) were generated by crossing 2afl/fl with Sftpc-CreERT2 driver (B6.129S-Sftpctm1(cre/ERT2)Blh/J; The Jackson Laboratory) (41). All mice were housed in colony cages on a 12 h light/12 h dark cycle with controlled humidity and temperature at 23 oC. All mice were fed ad libitum on a normal chow diet (Global 18% Protein Rodent Diet from Harlan, Envigo) and have free access to water. Pups were weaned at 3 weeks of age. Both male and female mice were used in all experiments.
Tamoxifen preparation and induction protocol
Tamoxifen (catalog no.: T5648-5G; Sigma) for injection was prepared by first dissolving powder in 100% ethanol. Corn oil was added to make a final concentration of 20 mg/ml, and ethanol was allowed to completely evaporate under nitrogen stream. Tamoxifen was filter-sterilized and stored at −20 °C. For reporter experiment, Mfsd2aERT2creTdTomato mice were administered with 250 μg/g body weight tamoxifen intraperitoneally for four consecutive days. For all experiments, AT2aKO and 2afl/fl controls were administered with 150 μg/g body weight tamoxifen intraperitoneally every other day for a total of three injections.
Antibodies
The following antibodies were used: Mfsd2a (in-house), Na+/K+ ATPase α1 (catalog no.: NB300-146; Novus Biologicals), pro-Sftpc (catalog no.: AB3786; Merck Millipore), Pdpn (catalog no.: MABT1512-100UG; Merck Millipore), F4/80 (catalog no.: ab6640; Abcam), RFP (catalog no.: 600-401-379; Rockland Immunochemicals), pro-Sftpb (catalog no.: AB3430; Merck Millipore), and Alexa Fluor secondary antibodies (Thermo Fisher Scientific).
Histological studies using paraffin sections
The lung was isolated from deeply anesthetized mice and fixed in 4% paraformaldehyde (PFA) in PBS at 4 °C overnight. After separating the lung into different lobes, the lobes were dehydrated in increasing concentrations of ethanol and xylene before embedding in paraffin. Lung sections (5 μm) were obtained using microtome (catalog no.: RM2255; Leica), and H&E staining was performed. Masson's trichome staining was performed to assess fibrosis in the lung. Images were obtained using BX53 Light Microscope (Olympus). In Figure 2C, AT2 cell diameters were quantified from four to seven images per mouse using ImageJ (64) and represented as individual points on a scatterplot. For immunofluorescence analysis, antigen retrieval was performed by boiling sections in sodium citrate buffer for 10 min in a microwave and allowed to cool to room temperature (RT). Lung sections were incubated with blocking buffer (10% normal goat serum, 1% bovine serum albumin [BSA] and 3 M glycine in Tris-buffered saline [TBS]–0.1% Triton-X [Tx]) for 1 h at RT. After blocking, antibodies were diluted in same blocking buffer without Tx and incubated overnight at 4 °C. The following antibodies were used: Mfsd2a (1:100 dilution), pro-Sftpc (1:200 dilution), Pdpn (1:200 dilution), and F4/80 (1:100 dilution). After washing with 0.1% Tx (TBS–Tx), lung sections were incubated with Alexa Fluor secondary antibodies (1:250) for 1 h at RT. Nuclei were stained with Hoechst 33342 (1:500 dilution; catalog no.: H3570; Thermo Fisher Scientific) for 5 min before mounting with FluorSave Reagent (catalog no.: 345789-20MLCN; Merck Millipore). Images were obtained using LSM710 Confocal Microscope (Carl Zeiss). In Figure 3B and Movie S1, 3D reconstruction was carried out on z-stack of 21 confocal images of lung section stained for Mfsd2a and the AT2 basolateral membrane marker Na+/K+ ATPase α1 using ZEN Digital Imaging for Light Microscopy software (Carl Zeiss).
Histological studies using frozen sections
The lung was isolated from deeply anesthetized mice and fixed in 4% PFA in PBS at 4 °C overnight. After separating the lung into different lobes, the lobes were rinsed briefly in PBS and cryoprotected in 30% sucrose in PBS overnight before embedding in optimum cutting temperature (Tissue-Tek; Sakura Finetek USA, Inc). Lung sections (20 μm) were obtained using Leica Cryostat CM1520. For immunofluorescence analysis, tissue sections were rinsed in PBS to remove optimum cutting temperature. Thereafter, tissue sections were incubated with blocking buffer (10% normal goat serum, 1% BSA, and 3 M glycine in TBS–0.1% Tx) for 1 h at RT. After blocking, antibodies were diluted in the same blocking buffer and incubated overnight at 4 °C. The following antibodies were used: RFP (1:100 dilution) and Pdpn (1:200 dilution). After washing with TBS–Tx, tissue sections were incubated with Alexa Fluor secondary antibodies (1:250 dilution) for 1 h at RT. Nuclei were stained with Hoechst 33342 for 5 min before mounting with FluorSave Reagent. Images were obtained using LSM710 Confocal Microscope. For Figs. S1B and S2B, mean fluorescence of Sftpc and Sftpb was quantified from two to three images per mouse using ImageJ and represented as individual points on a scatterplot.
AT2 cell harvest
AT2 cells were harvested from deeply anesthetized AT2aKO and 2afl/fl control mice as described (65) with modifications. Lungs were digested in 30 U/ml Dispase II (catalog no.: 17105041; Gibco) in Ca2+/Mg2+-free Hank’s balanced salt solution. CD45-negative cells were first enriched from whole lung cell suspension using magnetic separation on LS Columns (catalog no.: 130-04-401; Miltenyi Biotec) using CD45 MicroBeads (catalog no.: 130-052-301; Miltenyi Biotec). AT2 cells were purified using Biotinylated antimouse CD326/EpCAM antibody (catalog no.: 130-117-751; Miltenyi Biotec) and antibiotin MicroBeads (catalog no.: 130-090-485; Miltenyi Biotec) and used for LPC-NBD transport, lipidomics analysis, and RNA-Seq.
LPC-NBD transport
Fluorescently labeled LPC with a 16:1 fatty acyl chain (LPC-NBD) was a kind gift from Travecta Therapeutics. A 2 μg/μl of LPC-NBD stock solution was prepared in 12% FA-free BSA (catalog no.: A7030; Sigma), and LPC-NBD transport was accessed in both AT2aKO and 2afl/fl controls 2 weeks after tamoxifen induction. Intratracheal instillation was carried out as described (66) with modifications. A total of 60 μg of LPC-NBD was instilled directly into the trachea of both AT2aKO and 2afl/fl controls. After 2 h, 500 μl of 0.1% BSA was instilled into the lungs to rinse out any excess LPC-NBD in the alveoli. Lungs were harvested and prepared as described previously for frozen sections. For intravenous delivery, 200 μg of LPC-NBD was injected intravenously into both AT2aKO and 2afl/fl controls via the retro-orbital plexus. After 2 h, lungs were perfused with 0.1% BSA through the right ventricle of the heart to rinse out excess LPC-NBD in the blood. Lungs were harvested and prepared as described previously for frozen sections. The following antibodies were used: pro-Sftpc (1:100 dilution) and F4/80 (1:100 dilution). To access LPC-NBD in isolated AT2 cells, one million AT2 cells were freshly isolated from lungs of AT2aKO and 2afl/fl controls 2 weeks after tamoxifen induction and incubated with 30 μg of LPC-NBD for 30 min with rotation at RT. Cells were rinsed with 0.5% BSA to remove excess LPC-NBD and fixed in 4% PFA. Cells were stained with pro-Sftpc (1:200 dilution) for 1 h at RT. Nuclei were stained with Hoechst 33342 for 5 min. Images were obtained using LSM710 Confocal Microscope.
Red/Green BODIPY PC-A2 transport
To prepare hydrolyzed Red/Green BODIPY PC-A2 (catalog no.: A10072; Thermo Fisher Scientific) and Bis-BODIPY FL C11-PC (catalog no.: B7701; Thermo Fisher Scientific) to access uptake in cells, 100 units of PLA2 (catalog no.: P9279; Sigma) was added to 20 μg of fluorescent probe and allowed to hydrolyze for 30 min at RT. PLA2 hydrolyzed products were extracted with chloroform:methanol. After drying under argon stream, lipid film was resolubilized in 12% FA-free BSA. Human embryonic kidney 293 (American Type Culture Collection) cells were seeded onto an 8-well glass chamber slide at a density of 5 × 105 cells/well and allowed to adhere for 24 h prior to transfection. Cells were transfected with 70 ng WT Mfsd2a or transport-deficient D97A Mfsd2a per well. Transfected cells were washed once with charcoal-stripped serum-free Dulbecco's modified Eagle's medium (Gibco) and 50 μM of PLA2 hydrolyzed products were added to each well. Following a 20 min of incubation, cells were washed twice with 0.5% BSA in charcoal-stripped serum-free Dulbecco's modified Eagle's medium to remove unbound lipids before staining with Hoechst. Live cell images were obtained using LSM710 Confocal Microscope. To assess uptake of BODIPY PC-A2 in mice, a 0.33 μg/μl of stock solution was prepared in 12% BSA, and uptake was accessed in both AT2aKO and 2afl/fl controls 2 weeks after tamoxifen induction. A total of 10 μg of BODIPY PC-A2 was instilled directly into the trachea of both AT2aKO and 2afl/fl controls. After 2 h, 500 μl of 0.1% BSA was instilled into the lungs to rinse out any excess BODIPY PC-A2 in the alveoli. Lungs were harvested and prepared as described previously for frozen sections. The following antibodies were used: pro-Sftpc (1:100 dilution) and F4/80 (1:100 dilution). Nuclei were stained with Hoechst 33342 for 5 min. Images were obtained using LSM710 Confocal Microscope. Quantification of red and green fluorescence was carried out on 5 to 10 lung sections per mouse and represented as mean integrated density.
PLA A2 assay
PLA A2 (PLA2) activity assay was carried out on BAL isolated from AT2aKO and 2afl/fl control mice 2 weeks after tamoxifen induction. A short spin at 500g, 5 min at 4 °C, was carried out to remove cells. Red/Green BODIPY PC-A2 (Thermo Fisher Scientific; catalog no.: A10072) and Bis-BODIPY FL C11-PC (Thermo Fisher Scientific; catalog no.: B7701) were dissolved in ethanol to prepare a 20 μM stock. Red/Green BODIPY PC-A2 can be hydrolyzed by PLA2 to produce a red LPC and green FA. Bis-BODIPY FL C11-PC can be hydrolyzed by PLA1/2 to produce a green LPC and green FA. To prepare vesicles, 0.6 μM of substrate, 0.1% BSA, and 10 mM CaCl2 were added to 50 mM Tris–HCl, 100 mM NaCl, 1 mM EGTA (pH 7.5), and vortexed for 2 min. Vesicles were added directly to BAL and incubated for 1.5 h at 37 °C with shaking. Unhydrolyzed control was prepared by adding vesicles to an equal volume of PBS. Lipids were extracted using chloroform:methanol and resolved by thin layer chromatography using a chloroform:methanol:H2O (65:25:4) solvent system. Plates were scanned at 488 and 568 nm using Azure Sapphire system.
Lipidomic analysis
Sample preparation
BAL was harvested from deeply anesthetized AT2aKO and 2afl/fl control mice 2 and 4 weeks after tamoxifen induction as described (67) using Ca2+/Mg2+-free PBS and processed for lipidomics analysis. About 20 μl of surfactant was mixed with 180 μl of butanol:methanol (1:1, v/v) spiked with internal standards (ISs). The standards were purchased from Avanti Lipids and included acylcarnitine 16:0 D3, cholesterol ester 18:0 D6, dihydroceramide d18:0/08:0, ceramide d18:1/12:0, deoxyceramide m18:1/12:0, DAG 15:0/15:0, GM3 d18:1/18:0 D3, monohexosylceramide d18:1/12:0, dihexosylceramide d18:1/12:0, trihexosylceramide d18:1/18:0 D3, LPC 13:0, lysophosphatidylethanolamine 14:0, PC 13:0/13:0, plasmalogen PC 18:0/18:1 D9, phosphatidylethanolamine 17:0/17:0, plasmalogen phosphatidylethanolamine 18:0/18:1 D9, PG 17:0/17:0, PI 12:0/13:0, phosphatidylserine 17:0/17:0, sphingomyelin d18:1/12:0, sphingosine d18:1 D7, and triacylglycerol 12:0/12:0/12:0. The mixture was vortexed for 10 s, sonicated for 30 min, and then centrifuged at 4 °C (14,000g for 10 min). The supernatant fraction was collected for LC–MS/MS analysis. A pooled lipid extract was used as a quality control (QC) sample and injected on every five study sample. AT2 cells were harvested from AT2aKO and 2afl/fl control mice 2 weeks after tamoxifen induction as described previously. Cell pellets (1.5 × 106 cells/sample) were mixed with 500 μl of butanol:methanol (1:1, v/v) spiked with the same ISs used for the experiment with BAL. The mixture was vortexed for 10 s, sonicated for 30 min, and then centrifuged at 4 °C (14,000g for 10 min). The supernatant fraction was collected for LC–MS/MS analysis. A pooled lipid extract was used as a QC sample and injected on every five study sample. Data were normalized to protein concentration in each sample.
LC–MS/MS and data analysis
LC–MS/MS analysis was performed on an Agilent UHPLC 1290 Infinity II liquid chromatography system connected to an Agilent QqQ 6495C. An Agilent Zorbax RRHD Eclipse Plus C18 column (2.1 × 50 mm, 1.8 μm) was used for the reversed-phase LC separation. The mobile phases A (60% water and 40% acetonitrile with 10 mmol/l ammonium formate) and B (10% acetonitrile and 90% isopropanol with 10 mmol/l ammonium formate) were used for the chromatographic separation. The following gradient was applied: 0 to 2 min, 20 to 60% B; 2 to 12 min, 60 to 100% B; 12 to 14 min, 100% B; and 14.01 to 15.8 min, 20% B. The oven temperature was maintained at 40 °C. Flow rate was set at 0.4 ml/min, and the sample injection volume was 1 μl. The positive ionization spray voltage and nozzle voltage were set at 3000 and 1000 V, respectively. The drying gas and sheath gas temperatures were both maintained at 250 °C. The drying gas and sheath gas flow rates were 14 and 11 l/min, respectively. The nebulizer nitrogen gas flow rate was set at 35 psi. The iFunnel high and low pressure RFs were 150 and 60 V, respectively. Targeted analysis was performed in dynamic multiple reaction monitoring positive-ion mode. The acquired MS data were analyzed using Agilent MassHunter software, version B.08.00. The signal-to-noise ratios were calculated using the raw peak areas in study samples and processed blanks (PBLK). Lipids that had signal-to-noise ratio of <10 and CV >20% in the QC samples and did not show a linear behavior (R2 <0.8) in dilution curves were excluded from the analysis. ISs were used to normalize the raw peak areas in the corresponding lipid class. For BAL samples, concentrations were further normalized to volume. Endogenous species were quantified using one standard per lipid class; thus, our method can only deliver relative quantitation results. BAL lipidomics data used for Figures 7 and 8, A–C are available in Tables S1 and S2, respectively. AT2 lipid species were classified as PLs, sphingolipids, or neutral lipids and represented as mol% of lipid class. Data used for Figure 8, D–G are available in Table S3.
RNA-Seq
AT2 cells were harvested from AT2aKO and 2afl/fl control mice 2 weeks after tamoxifen induction as described previously. Total RNA was extracted using RNeasy Mini kit (Qiagen), and RNA concentration was quantified using Nanodrop. About 1 μg RNA per sample was used for library preparation using NEBNext Ultra TM RNA Library Prep Kit for Illumina (NEB) according to the manufacturer's instructions, sequenced on the Illumina HiSeq2000 platform, and analysis was performed using Partek Flow (version 10). Paired-end reads were aligned to mm10 genome using STAR alignment 2.7.3a and annotated using RefSeq. Features were filtered using recommended parameters and median ratio normalized. Differential gene expression analysis was performed using DESeq2. Filtered gene lists (genes with fold change ≥1.5, p < 0.05) were used to perform enrichment analysis on gene sets using Gene Ontology to identify significantly altered functional groups.
Protein analysis
BAL was harvested from deeply anesthetized AT2aKO and 2afl/fl control mice 4 weeks after tamoxifen induction as described (67) using Ca2+/Mg2+-free PBS. Following a short spin to remove cell debris, protein concentration was quantified using BCA assay kit (catalog no.: 23225; Thermo Fisher Scientific). BAL was resolved under nonreducing conditions on 4 to 20% gradient gel, transferred to nitrocellulose membrane, and blocked in 0.5% nonfat milk. Antibodies were diluted in 3% BSA in 50 mM Tris, 150 mM NaCl, and 0.1% Tween-20 and incubated with the membrane at 4 °C overnight. Membranes were incubated with IR dye-labeled secondary antibodies (LI-COR) diluted in 3% BSA in 50 mM Tris, 150 mM NaCl, and 0.1% Tween-20. Immunoblot signals were captured by ODYSSEY infrared imaging system (LI-COR).
Statistical analysis
Unpaired t test with Welch’s correction was used to calculate statistical differences of AT2aKO and 2afl/fl control AT2 cell diameter (Fig. 2C); in vivo and in vitro PLA2 activity (Figs. 5C and 6D); different lipid species in AT2aKO and 2afl/fl mice BAL and AT2 (Figs. 7 and 8); Sftpc fluorescence in AT2aKO, SftpcCre-ERT2, and 2afl/fl AT2 (Fig. S1B) and Sftpb fluorescence in AT2aKO and 2afl/fl AT2 (Fig. S2B). All graphs and statistical tests were carried out on GraphPad Prism 9 (GraphPad Software, Inc) and R 3.4.0. A p < 0.05 was considered to be significant.
Data availability
Data are available on Geo Profiles (www.ncbi.nlm.nih.gov), accession number GSE186170.
Supporting information
This article contains supporting information.
Conflict of interest
D. L. S. is a scientific founder and advisor of Travecta Therapeutics that has developed a drug delivery platform that uses MFSD2A transport. All other authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
B. H. W. and D. L. S. conceptualization; B. H. W., F. T., and D. L. S. methodology; B. H. W., D. M., and D. L. S. formal analysis; B. H. W., D. M., G. L. C., and D. L. G. investigation; M. R. W. and D. L. S. resources; B. H. W. and D. L. S. writing–original draft; B. H. W., F. T., M. R. W., and D. L. S. writing–review and editing; F. T. and M. R. W. supervision; M. R. W. and D. L. S. funding acquisition.
Funding and additional information
This study was supported by a grant from the National Research Foundation (NRF), Singapore (grant no.: NRF-NRFI2017-05; to D. L. S.) and by Khoo Postdoctoral Research Fellowship Awards (to B. H. W.). Work in SLING (M. R. W., F. T., and D. M.) is supported by grants from the National University of Singapore via the Life Sciences Institute, the National Research Foundation (grant no.: NRFSBP-P4), the NRF, and A∗STAR Industry Alignment Fund-Industry Collaboration Project (grant no.: I1901E0040), and the NRF2017_SISFP08.
Edited by Dennis Voelker
Supporting information
References
- 1.Schmidt R., Meier U., Yabut-Perez M., Walmrath D., Grimminger F., Seeger W., Günther A. Alteration of fatty acid profiles in different pulmonary surfactant phospholipids in acute respiratory distress syndrome and severe pneumonia. Am. J. Respir. Crit. Care Med. 2001;163:95–100. doi: 10.1164/ajrccm.163.1.9903029. [DOI] [PubMed] [Google Scholar]
- 2.Gunther A., Schmidt R., Nix F., Yabut-Perez M., Guth C., Rosseau S., Siebert C., Grimminger F., Morr H., Velcovsky H.G., Seeger W. Surfactant abnormalities in idiopathic pulmonary fibrosis, hypersensitivity pneumonitis and sarcoidosis. Eur. Respir. J. 1999;14:565–573. doi: 10.1034/j.1399-3003.1999.14c14.x. [DOI] [PubMed] [Google Scholar]
- 3.Agudelo C.W., Kumley B.K., Area-Gomez E., Xu Y., Dabo A.J., Geraghty P., Campos M., Foronjy R., Garcia-Arcos I. Decreased surfactant lipids correlate with lung function in chronic obstructive pulmonary disease (COPD) PLoS One. 2020;15 doi: 10.1371/journal.pone.0228279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zepp J.A., Morrisey E.E. Cellular crosstalk in the development and regeneration of the respiratory system. Nat. Rev. Mol. Cell Biol. 2019;20:551–566. doi: 10.1038/s41580-019-0141-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pérez-Gil J. Structure of pulmonary surfactant membranes and films: The role of proteins and lipid-protein interactions. Biochim. Biophys. Acta. 2008;1778:1676–1695. doi: 10.1016/j.bbamem.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Whitsett J.A., Weaver T.E. Hydrophobic surfactant proteins in lung function and disease. N. Engl. J. Med. 2002;347:2141–2148. doi: 10.1056/NEJMra022387. [DOI] [PubMed] [Google Scholar]
- 7.Karnati S., Garikapati V., Liebisch G., Van Veldhoven P.P., Spengler B., Schmitz G., Baumgart-Vogt E. Quantitative lipidomic analysis of mouse lung during postnatal development by electrospray ionization tandem mass spectrometry. PLoS One. 2018;13 doi: 10.1371/journal.pone.0203464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griese M., Kirmeier H.G., Liebisch G., Rauch D., Stückler F., Schmitz G., Zarbock R. Surfactant lipidomics in healthy children and childhood interstitial lung disease. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ban N., Matsumura Y., Sakai H., Takanezawa Y., Sasaki M., Arai H., Inagaki N. ABCA3 as a lipid transporter in pulmonary surfactant biogenesis. J. Biol. Chem. 2007;282:9628–9634. doi: 10.1074/jbc.M611767200. [DOI] [PubMed] [Google Scholar]
- 10.Yamano G., Funahashi H., Kawanami O., Zhao L.X., Ban N., Uchida Y., Morohoshi T., Ogawa J., Shioda S., Inagaki N. ABCA3 is a lamellar body membrane protein in human lung alveolar type II cells. FEBS Lett. 2001;508:221–225. doi: 10.1016/s0014-5793(01)03056-3. [DOI] [PubMed] [Google Scholar]
- 11.Grabner R., Meerbach W. Phagocytosis of surfactant by alveolar macrophages in vitro. Am. J. Physiol. 1991;261:L472–L477. doi: 10.1152/ajplung.1991.261.6.L472. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida M., Whitsett J.A. Interactions between pulmonary surfactant and alveolar macrophages in the pathogenesis of lung disease. Cell Mol. Biol. (Noisy-le-grand) 2004;50 Online Pub:OL639–OL648. [PubMed] [Google Scholar]
- 13.Bernhard W., Pynn C.J., Jaworski A., Rau G.A., Hohlfeld J.M., Freihorst J., Poets C.F., Stoll D., Postle A.D. Mass spectrometric analysis of surfactant metabolism in human volunteers using deuteriated choline. Am. J. Respir. Crit. Care Med. 2004;170:54–58. doi: 10.1164/rccm.200401-089OC. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs H., Jobe A., Ikegami M., Conaway D. The significance of reutilization of surfactant phosphatidylcholine. J. Biol. Chem. 1983;258:4159–4165. [PubMed] [Google Scholar]
- 15.Bridges J.P., Ikegami M., Brilli L.L., Chen X., Mason R.J., Shannon J.M. LPCAT1 regulates surfactant phospholipid synthesis and is required for transitioning to air breathing in mice. J. Clin. Invest. 2010;120:1736–1748. doi: 10.1172/JCI38061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harayama T., Eto M., Shindou H., Kita Y., Otsubo E., Hishikawa D., Ishii S., Sakimura K., Mishina M., Shimizu T. Lysophospholipid acyltransferases mediate phosphatidylcholine diversification to achieve the physical properties required in vivo. Cell Metab. 2014;20:295–305. doi: 10.1016/j.cmet.2014.05.019. [DOI] [PubMed] [Google Scholar]
- 17.Gross N.J., Barnes E., Narine K.R. Recycling of surfactant in black and beige mice: Pool sizes and kinetics. J. Appl. Physiol. (1985) 1988;64:2017–2025. doi: 10.1152/jappl.1988.64.5.2017. [DOI] [PubMed] [Google Scholar]
- 18.Wright J.R. Clearance and recycling of pulmonary surfactant. Am. J. Physiol. 1990;259:L1–12. doi: 10.1152/ajplung.1990.259.2.L1. [DOI] [PubMed] [Google Scholar]
- 19.Rindler T.N., Stockman C.A., Filuta A.L., Brown K.M., Snowball J.M., Zhou W., Veldhuizen R., Zink E.M., Dautel S.E., Clair G., Ansong C., Xu Y., Bridges J.P., Whitsett J.A. Alveolar injury and regeneration following deletion of ABCA3. JCI Insight. 2017;2 doi: 10.1172/jci.insight.97381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melton K.R., Nesslein L.L., Ikegami M., Tichelaar J.W., Clark J.C., Whitsett J.A., Weaver T.E. SP-B deficiency causes respiratory failure in adult mice. Am. J. Physiol. Lung Cell Mol. Physiol. 2003;285:L543–L549. doi: 10.1152/ajplung.00011.2003. [DOI] [PubMed] [Google Scholar]
- 21.Tian Y., Zhou R., Rehg J.E., Jackowski S. Role of phosphocholine cytidylyltransferase alpha in lung development. Mol. Cell. Biol. 2007;27:975–982. doi: 10.1128/MCB.01512-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen L.N., Ma D., Shui G., Wong P., Cazenave-Gassiot A., Zhang X., Wenk M.R., Goh E.L., Silver D.L. Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature. 2014;509:503–506. doi: 10.1038/nature13241. [DOI] [PubMed] [Google Scholar]
- 23.Wong B.H., Chan J.P., Cazenave-Gassiot A., Poh R.W., Foo J.C., Galam D.L., Ghosh S., Nguyen L.N., Barathi V.A., Yeo S.W., Luu C.D., Wenk M.R., Silver D.L. Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid (DHA) in eye and is important for photoreceptor cell development. J. Biol. Chem. 2016;291:10501–10514. doi: 10.1074/jbc.M116.721340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baisted D.J., Robinson B.S., Vance D.E. Albumin stimulates the release of lysophosphatidylcholine from cultured rat hepatocytes. Biochem. J. 1988;253:693–701. doi: 10.1042/bj2530693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan J.P., Wong B.H., Chin C.F., Galam D.L.A., Foo J.C., Wong L.C., Ghosh S., Wenk M.R., Cazenave-Gassiot A., Silver D.L. The lysolipid transporter Mfsd2a regulates lipogenesis in the developing brain. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2006443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobanova E.S., Schuhmann K., Finkelstein S., Lewis T.R., Cady M.A., Hao Y., Keuthan C., Ash J.D., Burns M.E., Shevchenko A., Arshavsky V.Y. Disrupted blood-retina lysophosphatidylcholine transport impairs photoreceptor health but not visual signal transduction. J. Neurosci. 2019;39:9689–9701. doi: 10.1523/JNEUROSCI.1142-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreone B.J., Chow B.W., Tata A., Lacoste B., Ben-Zvi A., Bullock K., Deik A.A., Ginty D.D., Clish C.B., Gu C. Blood-brain barrier permeability is regulated by lipid transport-dependent suppression of caveolae-mediated transcytosis. Neuron. 2017;94:581–594.e5. doi: 10.1016/j.neuron.2017.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guemez-Gamboa A., Nguyen L.N., Yang H., Zaki M.S., Kara M., Ben-Omran T., Akizu N., Rosti R.O., Rosti B., Scott E., Schroth J., Copeland B., Vaux K.K., Cazenave-Gassiot A., Quek D.Q., et al. Inactivating mutations in MFSD2A, required for omega-3 fatty acid transport in brain, cause a lethal microcephaly syndrome. Nat. Genet. 2015;47:809–813. doi: 10.1038/ng.3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alakbarzade V., Hameed A., Quek D.Q., Chioza B.A., Baple E.L., Cazenave-Gassiot A., Nguyen L.N., Wenk M.R., Ahmad A.Q., Sreekantan-Nair A., Weedon M.N., Rich P., Patton M.A., Warner T.T., Silver D.L., et al. A partially inactivating mutation in the sodium-dependent lysophosphatidylcholine transporter MFSD2A causes a non-lethal microcephaly syndrome. Nat. Genet. 2015;47:814–817. doi: 10.1038/ng.3313. [DOI] [PubMed] [Google Scholar]
- 30.Harel T., Quek D.Q.Y., Wong B.H., Cazenave-Gassiot A., Wenk M.R., Fan H., Berger I., Shmueli D., Shaag A., Silver D.L., Elpeleg O., Edvardson S. Homozygous mutation in MFSD2A, encoding a lysolipid transporter for docosahexanoic acid, is associated with microcephaly and hypomyelination. Neurogenetics. 2018;19:227–235. doi: 10.1007/s10048-018-0556-6. [DOI] [PubMed] [Google Scholar]
- 31.Scala M., Chua G.L., Chin C.F., Alsaif H.S., Borovikov A., Riazuddin S., Riazuddin S., Chiara Manzini M., Severino M., Kuk A., Fan H., Jamshidi Y., Toosi M.B., Doosti M., Karimiani E.G., et al. Biallelic MFSD2A variants associated with congenital microcephaly, developmental delay, and recognizable neuroimaging features. Eur. J. Hum. Genet. 2020;28:1509–1519. doi: 10.1038/s41431-020-0669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quek D.Q., Nguyen L.N., Fan H., Silver D.L. Structural insights into the transport mechanism of the human sodium-dependent lysophosphatidylcholine transporter Mfsd2a. J. Biol. Chem. 2016;291:9383–9394. doi: 10.1074/jbc.M116.721035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berger J.H., Charron M.J., Silver D.L. Major facilitator superfamily domain-containing protein 2a (MFSD2A) has roles in body growth, motor function, and lipid metabolism. PLoS One. 2012;7 doi: 10.1371/journal.pone.0050629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piccirillo A.R., Hyzny E.J., Beppu L.Y., Menk A.V., Wallace C.T., Hawse W.F., Buechel H.M., Wong B.H., Foo J.C., Cazenave-Gassiot A., Wenk M.R., Delgoffe G.M., Watkins S.C., Silver D.L., D'Cruz L.M. The lysophosphatidylcholine transporter MFSD2A is essential for CD8(+) memory T cell maintenance and secondary response to infection. J. Immunol. 2019;203:117–126. doi: 10.4049/jimmunol.1801585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mae M.A., He L., Nordling S., Vazquez-Liebanas E., Nahar K., Jung B., Li X., Tan B.C., Chin Foo J., Cazenave-Gassiot A., Wenk M.R., Zarb Y., Lavina B., Quaggin S.E., Jeansson M., et al. Single-cell analysis of blood-brain barrier response to pericyte loss. Circ. Res. 2021;128:e46–e62. doi: 10.1161/CIRCRESAHA.120.317473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adams T.S., Schupp J.C., Poli S., Ayaub E.A., Neumark N., Ahangari F., Chu S.G., Raby B.A., DeIuliis G., Januszyk M., Duan Q., Arnett H.A., Siddiqui A., Washko G.R., Homer R., et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Habermann A.C., Gutierrez A.J., Bui L.T., Yahn S.L., Winters N.I., Calvi C.L., Peter L., Chung M.I., Taylor C.J., Jetter C., Raju L., Roberson J., Ding G., Wood L., Sucre J.M.S., et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McDonough J.E., Ahangari F., Li Q., Jain S., Verleden S.E., Herazo-Maya J., Vukmirovic M., DeIuliis G., Tzouvelekis A., Tanabe N., Chu F., Yan X., Verschakelen J., Homer R.J., Manatakis D.V., et al. Transcriptional regulatory model of fibrosis progression in the human lung. JCI Insight. 2019;4 doi: 10.1172/jci.insight.131597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koenitzer J.R., Wu H., Atkinson J.J., Brody S.L., Humphreys B.D. Single-nucleus RNA-sequencing profiling of mouse lung. Reduced dissociation bias and improved rare cell-type detection compared with single-cell RNA sequencing. Am. J. Respir. Cell Mol. Biol. 2020;63:739–747. doi: 10.1165/rcmb.2020-0095MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cater R.J., Chua G.L., Erramilli S.K., Keener J.E., Choy B.C., Tokarz P., Chin C.F., Quek D.Q.Y., Kloss B., Pepe J.G., Parisi G., Wong B.H., Clarke O.B., Marty M.T., Kossiakoff A.A., et al. Structural basis of omega-3 fatty acid transport across the blood-brain barrier. Nature. 2021;595:315–319. doi: 10.1038/s41586-021-03650-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rock J.R., Barkauskas C.E., Cronce M.J., Xue Y., Harris J.R., Liang J., Noble P.W., Hogan B.L. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc. Natl. Acad. Sci. U. S. A. 2011;108:E1475–E1483. doi: 10.1073/pnas.1117988108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glasser S.W., Burhans M.S., Korfhagen T.R., Na C.L., Sly P.D., Ross G.F., Ikegami M., Whitsett J.A. Altered stability of pulmonary surfactant in SP-C-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6366–6371. doi: 10.1073/pnas.101500298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arbibe L., Koumanov K., Vial D., Rougeot C., Faure G., Havet N., Longacre S., Vargaftig B.B., Béréziat G., Voelker D.R., Wolf C., Touqui L. Generation of lyso-phospholipids from surfactant in acute lung injury is mediated by type-II phospholipase A2 and inhibited by a direct surfactant protein A-phospholipase A2 protein interaction. J. Clin. Invest. 1998;102:1152–1160. doi: 10.1172/JCI3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Madsen J., Panchal M.H., Mackay R.A., Echaide M., Koster G., Aquino G., Pelizzi N., Perez-Gil J., Salomone F., Clark H.W., Postle A.D. Metabolism of a synthetic compared with a natural therapeutic pulmonary surfactant in adult mice. J. Lipid Res. 2018;59:1880–1892. doi: 10.1194/jlr.M085431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scarpelli E.M., Condorelli S., Colacicco G., Cosmi E.V. Lamb fetal pulmonary fluid. II. Fate of phosphatidylcholine. Pediatr. Res. 1975;9:195–201. doi: 10.1203/00006450-197504000-00011. [DOI] [PubMed] [Google Scholar]
- 46.Oyarzun M.J., Clements J.A., Baritussio A. Ventilation enhances pulmonary alveolar clearance of radioactive dipalmitoyl phosphatidylcholine in liposomes. Am. Rev. Respir. Dis. 1980;121:709–721. doi: 10.1164/arrd.1980.121.4.709. [DOI] [PubMed] [Google Scholar]
- 47.Burdge G.C., Kelly F.J., Postle A.D. Synthesis of phosphatidylcholine in Guinea-pig fetal lung involves acyl remodelling and differential turnover of individual molecular species. Biochim. Biophys. Acta. 1993;1166:251–257. doi: 10.1016/0005-2760(93)90105-i. [DOI] [PubMed] [Google Scholar]
- 48.Post M., Schuurmans E.A., Batenburg J.J., Van Golde L.M. Mechanisms involved in the synthesis of disaturated phosphatidylcholine by alveolar type II cells isolated from adult rat lung. Biochim. Biophys. Acta. 1983;750:68–77. doi: 10.1016/0005-2760(83)90205-9. [DOI] [PubMed] [Google Scholar]
- 49.Martini W.Z., Chinkes D.L., Barrow R.E., Murphey E.D., Wolfe R.R. Lung surfactant kinetics in conscious pigs. Am. J. Physiol. 1999;277:E187–E195. doi: 10.1152/ajpendo.1999.277.1.E187. [DOI] [PubMed] [Google Scholar]
- 50.Hallman M., Epstein B.L., Gluck L. Analysis of labeling and clearance of lung surfactant phospholipids in rabbit. Evidence of bidirectional surfactant flux between lamellar bodies and alveolar lavage. J. Clin. Invest. 1981;68:742–751. doi: 10.1172/JCI110310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Numata M., Chu H.W., Dakhama A., Voelker D.R. Pulmonary surfactant phosphatidylglycerol inhibits respiratory syncytial virus-induced inflammation and infection. Proc. Natl. Acad. Sci. U. S. A. 2010;107:320–325. doi: 10.1073/pnas.0909361107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Numata M., Kandasamy P., Nagashima Y., Posey J., Hartshorn K., Woodland D., Voelker D.R. Phosphatidylglycerol suppresses influenza A virus infection. Am. J. Respir. Cell Mol. Biol. 2012;46:479–487. doi: 10.1165/rcmb.2011-0194OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Numata M., Grinkova Y.V., Mitchell J.R., Chu H.W., Sligar S.G., Voelker D.R. Nanodiscs as a therapeutic delivery agent: Inhibition of respiratory syncytial virus infection in the lung. Int. J. Nanomedicine. 2013;8:1417–1427. doi: 10.2147/IJN.S39888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuronuma K., Mitsuzawa H., Takeda K., Nishitani C., Chan E.D., Kuroki Y., Nakamura M., Voelker D.R. Anionic pulmonary surfactant phospholipids inhibit inflammatory responses from alveolar macrophages and U937 cells by binding the lipopolysaccharide-interacting proteins CD14 and MD-2. J. Biol. Chem. 2009;284:25488–25500. doi: 10.1074/jbc.M109.040832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Numata M., Kandasamy P., Nagashima Y., Fickes R., Murphy R.C., Voelker D.R. Phosphatidylinositol inhibits respiratory syncytial virus infection. J. Lipid Res. 2015;56:578–587. doi: 10.1194/jlr.M055723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kandasamy P., Numata M., Berry K.Z., Fickes R., Leslie C.C., Murphy R.C., Voelker D.R. Structural analogs of pulmonary surfactant phosphatidylglycerol inhibit toll-like receptor 2 and 4 signaling. J. Lipid Res. 2016;57:993–1005. doi: 10.1194/jlr.M065201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richmond B.L., Hui D.Y. Molecular structure and tissue-specific expression of the mouse pancreatic phospholipase A(2) gene. Gene. 2000;244:65–72. doi: 10.1016/s0378-1119(00)00006-8. [DOI] [PubMed] [Google Scholar]
- 58.Hallstrand T.S., Chi E.Y., Singer A.G., Gelb M.H., Henderson W.R. Secreted phospholipase A2 group X overexpression in asthma and bronchial hyperresponsiveness. Am. J. Respir. Crit. Care Med. 2007;176:1072–1078. doi: 10.1164/rccm.200707-1088OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Munoz N.M., Meliton A.Y., Meliton L.N., Dudek S.M., Leff A.R. Secretory group V phospholipase A2 regulates acute lung injury and neutrophilic inflammation caused by LPS in mice. Am. J. Physiol. Lung Cell Mol. Physiol. 2009;296:L879–L887. doi: 10.1152/ajplung.90580.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mouchlis V.D., Bucher D., McCammon J.A., Dennis E.A. Membranes serve as allosteric activators of phospholipase A2, enabling it to extract, bind, and hydrolyze phospholipid substrates. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E516–E525. doi: 10.1073/pnas.1424651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carman G.M., Deems R.A., Dennis E.A. Lipid signaling enzymes and surface dilution kinetics. J. Biol. Chem. 1995;270:18711–18714. doi: 10.1074/jbc.270.32.18711. [DOI] [PubMed] [Google Scholar]
- 62.Gelb M.H., Jain M.K., Hanel A.M., Berg O.G. Interfacial enzymology of glycerolipid hydrolases: Lessons from secreted phospholipases A2. Annu. Rev. Biochem. 1995;64:653–688. doi: 10.1146/annurev.bi.64.070195.003253. [DOI] [PubMed] [Google Scholar]
- 63.Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., Lein E.S., Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sinha M., Lowell C.A. Isolation of highly pure primary mouse alveolar epithelial type II cells by flow cytometric cell sorting. Bio Protoc. 2016;6 doi: 10.21769/BioProtoc.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Helms M.N., Torres-Gonzalez E., Goodson P., Rojas M. Direct tracheal instillation of solutes into mouse lung. J. Vis. Exp. 2010 doi: 10.3791/1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang R., Zhang Y., Qian W., Peng L., Lin L., Xu J., Xie T., Ji J., Zhan X., Shan J. Surfactant lipidomics of alveolar lavage fluid in mice based on ultra-high-performance liquid chromatography coupled to hybrid quadrupole-exactive orbitrap mass spectrometry. Metabolites. 2019;9:80. doi: 10.3390/metabo9040080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available on Geo Profiles (www.ncbi.nlm.nih.gov), accession number GSE186170.