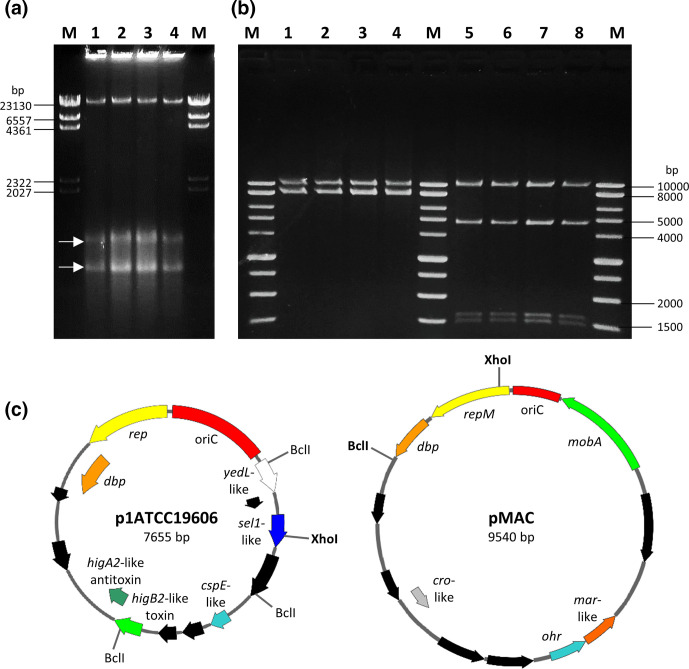
Fig. 5.
Plasmids p1ATCC19606 and pMAC harboured by A. baumannii ATCC 19606T strains. (a) Agarose gel electrophoresis of clear lysates of A. baumannii ATCC 19606(A) (lane 1), ATCC 19606(D) (lane 2), ATCC 19606(S) (lane 3) and ATCC 19606(T) (lane 4). M, Lambda DNA/HindIII marker (ThermoFisher). White arrows indicate the closed circular forms of pMAC (upper band) and p1ATCC19606 (lower band). (b) p1ATCC19606 and pMAC were copurified from A. baumannii strains ATCC 19606(A) (lanes 1 and 5), ATCC 19606(D) (lanes 2 and 6), ATCC 19606(S) (lanes 3 and 7) and ATCC 19606(T) (lanes 4 and 8), and digested with XhoI (lanes 1–4) and BclI (lanes 5–8). M, BenchTop 1 kb DNA Ladder (Promega). (c) Physical and functional maps of the p1ATCC19606 and pMAC plasmids. Restriction sites for the enzymes used to generate the electropherogram in (b) are shown. Unique cutter restriction enzymes are indicated in bold. Nomenclature of p1ATCC19606: rep, putative replicase; dbp, gene encoding a predicted DNA-binding protein; cspE-like, putative cold-shock protein gene; sel1-like, putative gene coding for a Sel1-repeat family protein; yedL-like, gene coding for the putative YedL N-acetyltransferase; oriC, predicted origin of replication. Nomenclature of pMAC: repM, replication protein M; dbp, gene encoding a predicted DNA-binding protein; ohr, gene encoding an organic hydroperoxide resistance protein, mobA, plasmid mobilization protein; oriC, origin of replication. ORFs shown in black are predicted to encode for hypothetical proteins. All genes are reported in scale over the total length of each plasmid. Images were obtained by the use of the SnapGene software (GSL Biotech).