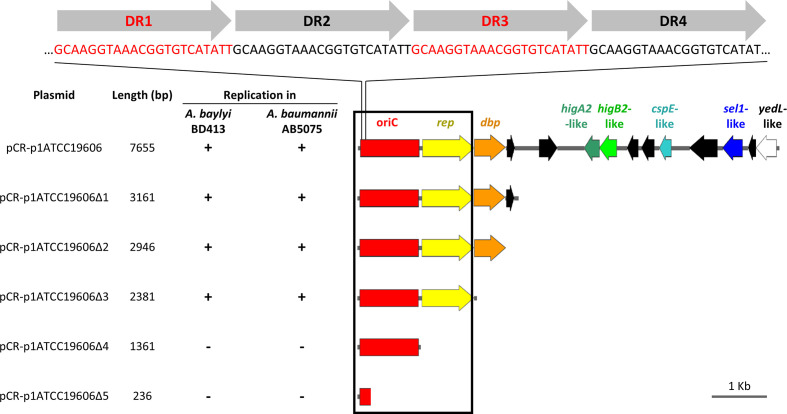
Fig. 6.
Deletion analysis of p1ATCC19606 to determine the minimal region required for autonomous plasmid replication in Acinetobacter spp. Deletion fragments of p1ATCC19606 were generated by PCR amplification with primers listed in Table S1 and cloned into pCR. The resulting p1ATCC19606 deletion derivatives were introduced in A. baylyi BD413 and A. baumannii AB5075 to map the minimal self-replicating region (black box). Relevant coding regions are indicated with colours: red, predicted minimal origin of replication (oriC); yellow, putative replicase (rep); orange, gene encoding a predicted DNA-binding protein (dbp); dark green, putative higA2-like antitoxin gene; light green, putative higB2-like toxin gene; cyan, putative cold-shock protein gene (cspE); blue, gene coding for putative a Sel1-repeat family protein (sel1); white, gene coding for the putative YedL N-acetyltransferase (yedL). Four copies of the 22-mer direct repeat (DR1–DR4) in the predicted origin of replication are shown on top. ORFs in black are predicted to encode for hypothetical proteins. All genes are reported in scale over the total length of the plasmid. Images were obtained by the use of the SnapGene software (GSL Biotech).