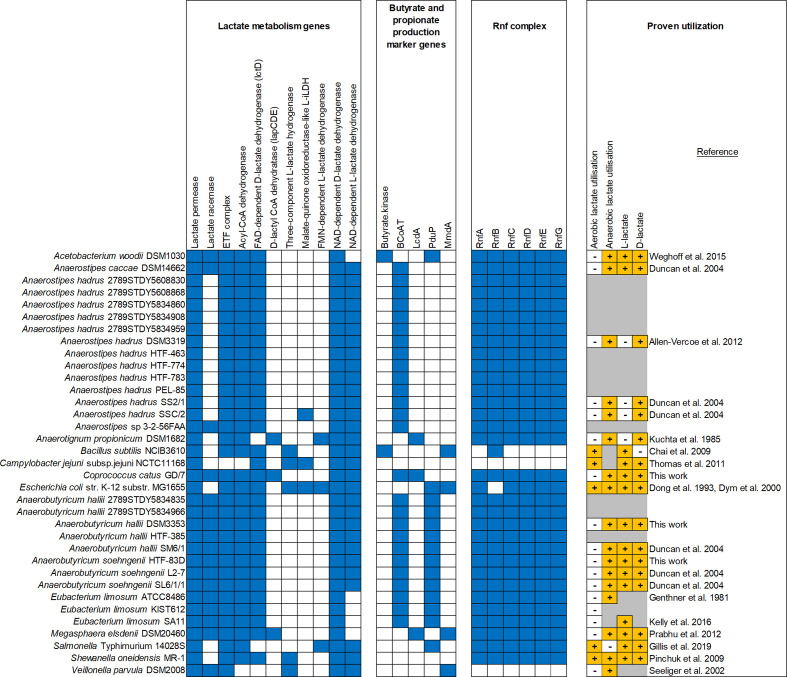
Fig. 5.
Distribution of lactate utilization and associated genes in lactate-utilizing bacteria. Presence (blue) and absence (white) of genes was determined as described in Methods. The ability (+) or inabilty (-) of selected strains to utilize different isomers of lactate during aerobic or anaerobic growth was obtained from the literature [12, 21–23, 57, 59, 67, 75–81]. Certain details could not be recovered for some strains (marked in grey). Full names of genes indicated by short-hand gene nomenclature are as follows: Butyryl-CoA:acetate CoA-transferase (BCoAT), lactoyl-CoA dehydratase, alpha subunit (LcdA), propanediol utilization CoA-dependent propionaldehyde dehydrogenase (PduP), methyl-malonyl-CoA decarboxylase, alpha-subunit (MmdA) and Rnf complex subunits (RnfA-G).