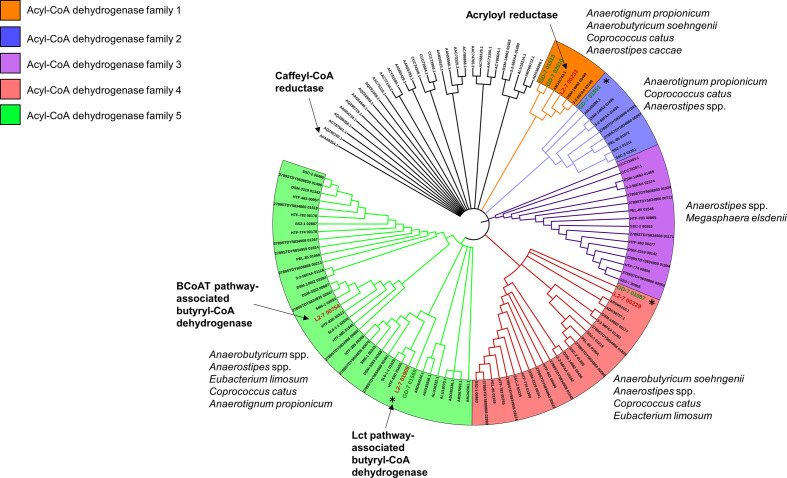
Fig. 7.
Phylogeny of Acyl-CoA dehydrogenase genes present in lactate-utilizing bacteria. Branch validation was performed using 1000 ultrafast bootstrap replicates and a hill-climbing nearest-neighbour interchange (NNI) search was performed to reduce the risk of overestimating branch supports. Branches with less than 90% UFBoot support were collapsed. Genes labelled ‘Caffeyl-CoA reductase’ and ‘Acryloyl reductase’ were experimentally verified by Bertsch et al. [61] and Hetzel et al. [62], respectively, and the gene labelled ‘Butyryl-CoA dehydrogenase’ is present in the butyrate production loci as described previously [58]. This maximum-likelihood tree was reconstructed using the LG+G4 model. The tree was rooted using minimal ancestor deviation. Anaerobutyricum soehngenii L2-7 and Coprococcus catus GD/7 genes are represented in red and green font, respectively. Asterisks indicate genes that were highly upregulated by lactate in the transcriptomics work presented in this study.