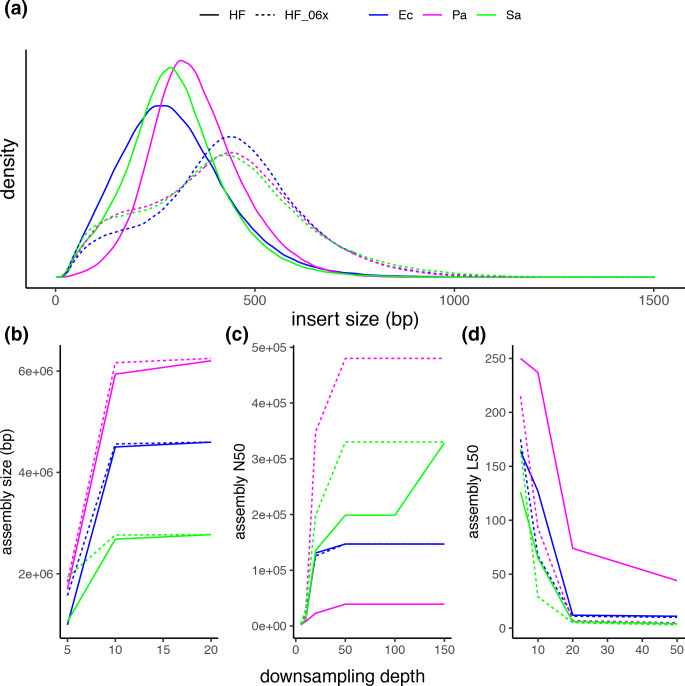
Fig. 4.
Insert size distribution and assembly metrics of double-left clean-up Hackflex libraries compared to Hackflex libraries. Insert size distributions (a) are shown for Hackflex and double-left clean-up Hackflex libraries from E. coli (blue) (Ec.HF.B3 median=287, mean=300.68; Ec.HF_06x.B3 median=433, mean=432.72), P. aeruginosa (pink) (Pa.HF.B2 median=341, mean=352.01; Pa.HF_06x.B3 median=417, mean=415.06) and S. aureus (green) (Sa.HF.B2 median=296, mean=304.94; Sa.HF_06x.B3 median=416, mean=420.29) genomic DNA. The same libraries, down sampled to several depths, and assembled, were analysed for assembly quality. Assembly size (b), and assembly metrics N50 (c) and L50 (d) are shown for Hackflex and double-left clean-up Hackflex libraries from E. coli (blue), P. aeruginosa (pink) and S. aureus (green).