FIGURE 1.
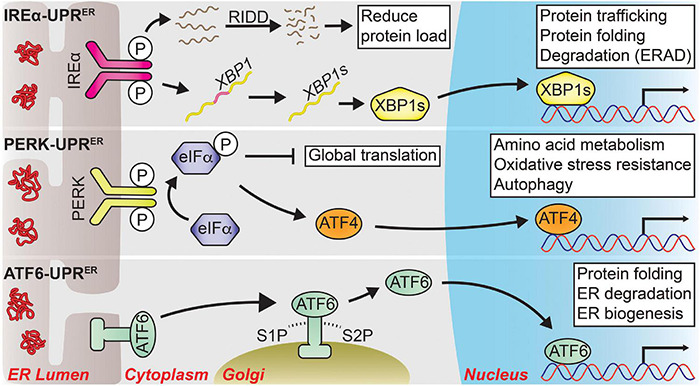
The three major signal transduction pathways of the UPRER. Following ER stress, three distinct branches are activated that shape the UPRER. IRE1α-UPRER: Once activated via its dimerization and autophosphorylation, IRE1α cleaves a select group of mRNAs and miRNAs to drive their degradation through a process known as regulated IRE1-dependent decay (RIDD), reducing the total protein folding load on the ER. IRE1α also facilitates the unconventional splicing of XBP1 mRNA into its spliced form, a potent transcription factor known as XBP1s, which drives the expression of genes tied to protein quality control to restore ER homeostasis. PERK-UPRER: PERK also dimerizes and autophosphorylates upon ER stress, which then phosphorylates eIF2α to attenuate global translation. The mRNA of transcription factor ATF4 is preferentially translated following eIF2α phosphorylation, allowing it to upregulate genes involved in amino acid metabolism, oxidative stress resistance, autophagy, and apoptosis. ATF6-UPRER: ER stress unmasks several Golgi-localization signals within ATF6 that allow it to translocate to the Golgi body. There, it is sequentially cleaved by site-1 protease (S1P) and site-2 protease (S2P) from its full-length form (ATF6p90) into its transcriptionally active form (ATF6p50), which initiates the transcription of UPR target genes pertaining to protein quality control and ER biogenesis to promote ER secretory capacity. Solid arrows represent direct actions.
