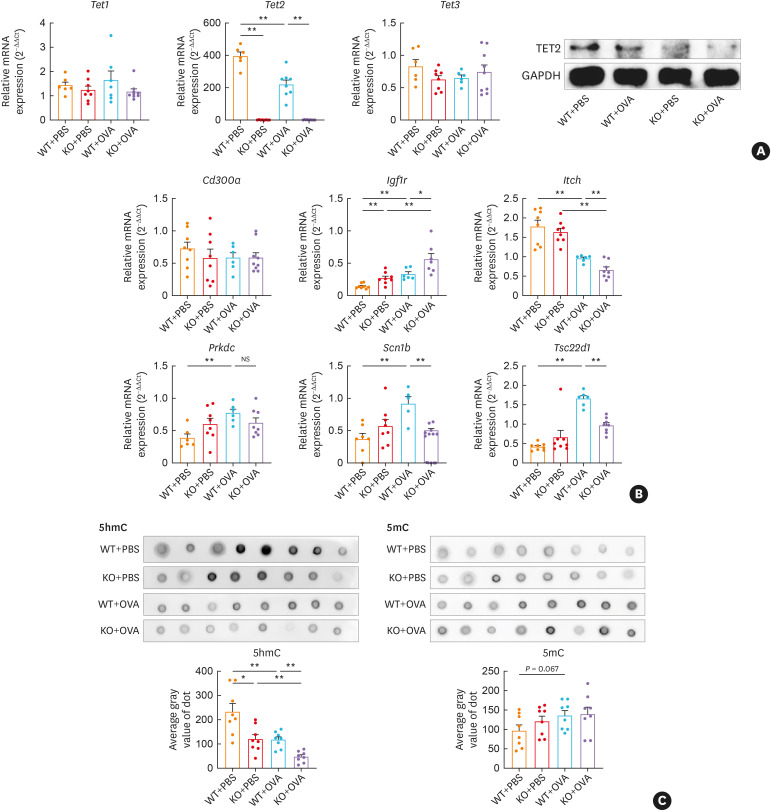
Fig. 6. Tet2 modulates DhMG expressions and DNA methylation in CD4+ T cells of OVA-exposed mice. (A) Relative mRNA expressions of Tet1, Tet2, and Tet3 in CD4+ T cells were determined by RT-qPCR and calculated using comparative Ct values after normalizing to β-actin. The relative protein levels were quantified by western blotting after normalizing to GAPDH. (B) Relative mRNA expressions of Cd300a, Igf1r, Itch, Prkdc, Scn1b, and Tsc22d1 in CD4+ T cells were determined by RT-qPCR and calculated using comparative Ct values after normalizing to β-actin. (C, D) Global DNA 5hmC deposition in mice CD4+ T cells was detected using dot blot and quantified using the average gray value of dot.
Tet, ten-eleven translocation; DhMG, differentially hydroxymethylated genes; PBS, phosphate-buffered saline; OVA, ovalbumin; WT, wild type; KO, Tet2 knockout; RT-qPCR, reverse real-time quantitative transcription polymerase chain reaction; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; Igf1r, insulin-like growth factor 1 receptor; Itch, itchy E3 ubiquitin protein ligase; Prkdc, protein kinase, DNA-activated, catalytic subunit; Scn1b, sodium voltage-gated channel beta subunit 1; Tsc22d1, transforming growth factor β-stimulated clone 22 domain family member 1; NS, not significant.
*P < 0.05, **P < 0.01.