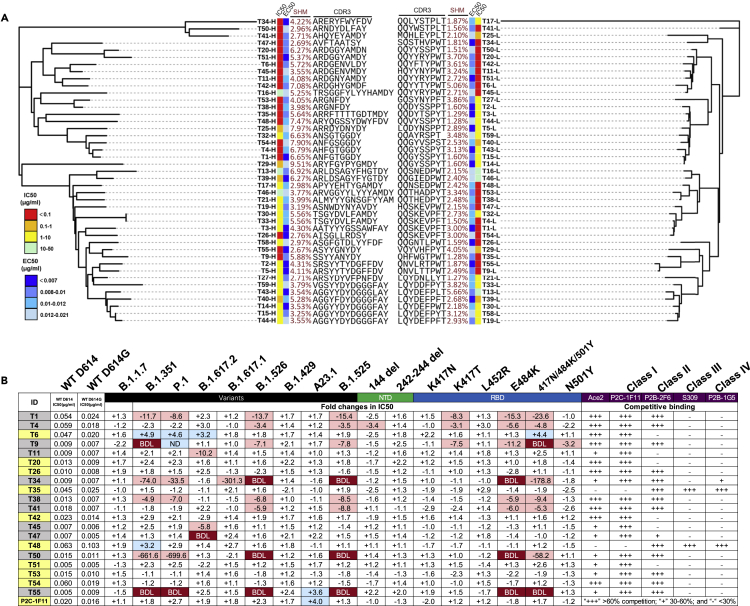
Figure 5.
RBD trimer induces dominant broad and potent monoclonal antibodies against SARS-CoV-2 variants
(A) Unrooted neighbor-joining tree depicting the genetic relationship among 40 monoclonal antibodies. The left panel is derived from the heavy chain variable region while the right panel from the light chain variable region. Antibody names, and their corresponding IC50, EC50, degree of somatic hyper mutation, and CDR3 sequences are indicated at the tip of the branches. The IC50 was measured against SARS-CoV-2 WT D614 pseudovirus and the EC50 was calculated against recombinant RBD trimer in the ELISA assay.
(B) IC50 and their fold changes against SARS-CoV-2 variants, as well as single or triple mutant pseudoviruses, relative to that of WT D614G. The symbol “+” indicates increased while “-” decreased potency. Those highlighted in blue indicates increased potency at least threefold whereas those in red decreased potency at least threefold. Those in white change less than threefold. BDL, referring to Below Detection Limit, indicates the highest concentration (2 μg/mL) of mAbs failed to reach 50% neutralization. Results were calculated from three independent experiments. Epitope mapping through competitive binding with ACE2 and various well-defined monoclonal antibodies (P2C-1F11, P2B-2F6, S309, and P2B-1G5) was also shown. “+++” indicates >60% competition, while “+” 30%–60%, and “-” <30%. Results are representatives of two independent experiments.