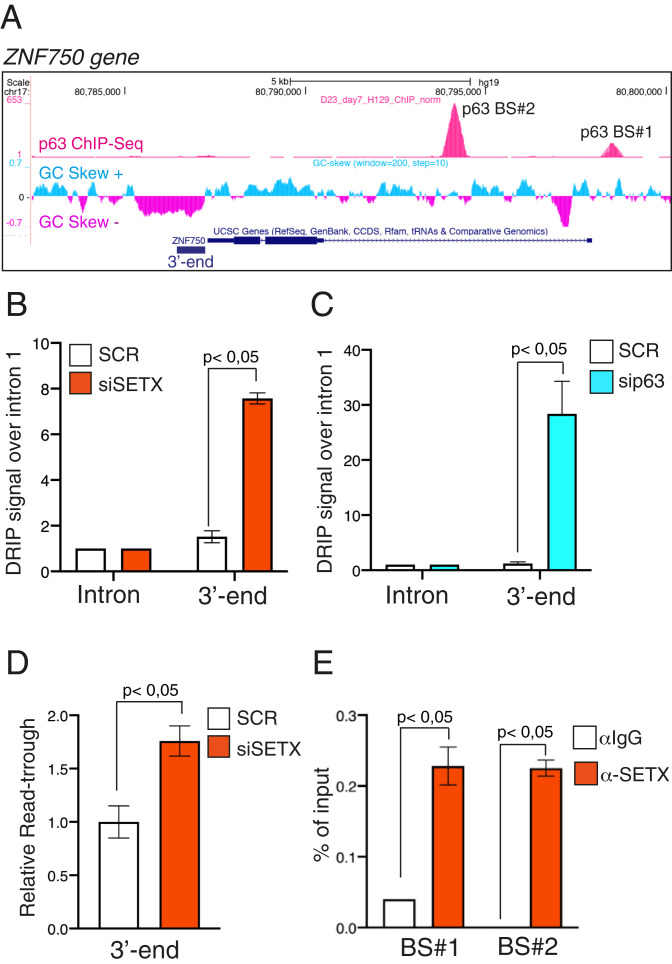
Fig. 3.
SETX regulates R-loop formation on the 3′ end of the p63 target gene ZNF750. (A) Schematic representation of ZNF750 genetic locus (modified from Genome Browser, https://genome.ucsc.edu) showing from top to bottom p63 ChIP-seq peaks in human keratinocytes (12), GC skew signal, and gene structure with exons (boxes) and introns (lines). (B) Quantification by qPCR of DRIP signal in differentiated keratinocytes treated with SCR or SETX siRNAs. Values at 3′ ends were normalized to ZNF750 intron 1 signal. Bars represent the mean of three replicates (n = 3, PCR runs) ± SD and are representative of three independent experiments (n = 3 biological replicates). P value was calculated by Student’s t test. (C) Quantification by qPCR of DRIP signal in differentiated keratinocytes treated with SCR or p63 siRNAs. Values at 3′ ends were normalized to ZNF750 intron 1 signal. Bars represent the mean of three replicates (n = 3, PCR runs) ± SD and are representative of two independent experiments (n = 2 biological replicates). P value was calculated by Student’s t test. (D) RT-qPCR analysis of readthrough transcripts at the ZNF750 3′ region in SCR or siSETX differentiated keratinocytes. Values are normalized to ZNF750 intron 1 and showed as fold enrichment of siSETX over SCR. Bars represent the mean of three replicates (n = 3, PCR runs) ± SD and are representative of three independent experiments (n = 3 biological replicates). P value was calculated by Student’s t test. (E) Quantification by qPCR of ChIP analysis of endogenous SETX occupancy at p63-binding sites (BS#1 and BS#2) in differentiated human keratinocytes. Bars represent the mean of three replicates (n = 3, PCR runs) ± SD and are representative of three independent experiments (n = 3 biological replicates). P value was calculated by Student’s t test.