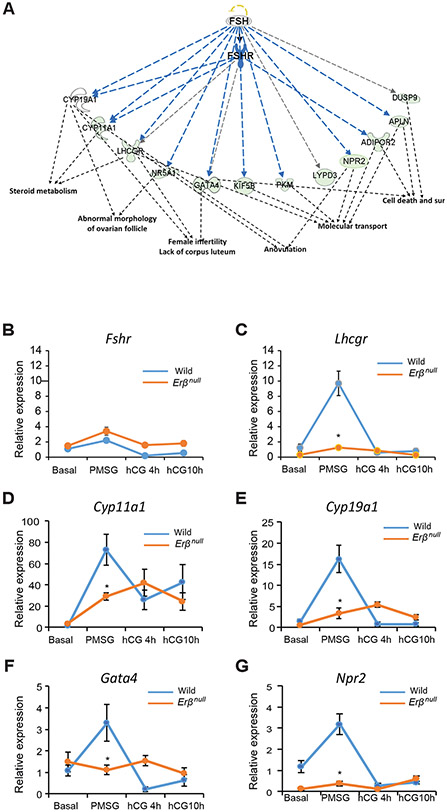
Figure 4. PMSG-regulated genes that are ERβ dependent and involved in folliculogenesis.
PMSG and ERβ dependent target genes that were identified in the Fig. 3A were subjected to Ingenuity Pathway Analysis (IPA). IPA revealed a group of downregulated genes that have FSH/FSHR as an upstream regulator (A) and are involved in steroid metabolism, folliculogenesis, ovulation, molecular transport, cell death, and cell survival (sur). The expression of these genes was further validated by RT-qPCR analysis (B-G). We did not detect any significant difference in the expression levels or pattern of Fshr between wildtype and Erβnull GCs (B). In wildtype GCs, Lhcgr, Cyp11a1 Cyp19a1, Gata4 and Npr2 expression significantly increased from Basal to PMSG and then returned to basal levels at 4h after hCG and remained unchanged at 10h after hCG (C-G). In Erβnull GCs the pattern of expression of Lhcgr was similar to that of wildtype GCs, but following PMSG Lhcgr expression was significantly lower in Erβnull GCs compared to wildtype GCs (C). Cyp11a1 and Cyp19a1 expression showed an increasing pattern from Basal to hCG4h and remained unchanged at hCG10h, however, following PMSG Cyp11a1 and Cyp19a1 expression was significantly lower in Erβnull GCs compared to wildtype GCs (D, E). There was no significant change in the expression of Gata4 and Npr2 from Basal to 10h hCG. But following PMSG treatment, Gata4 and Npr2 expression was significantly lower in Erβnull GCs compared to wildtype GCs (F, G). RT-qPCR data are represented as mean ± SEM. n ≥ 6. *P ≤ 0.05.