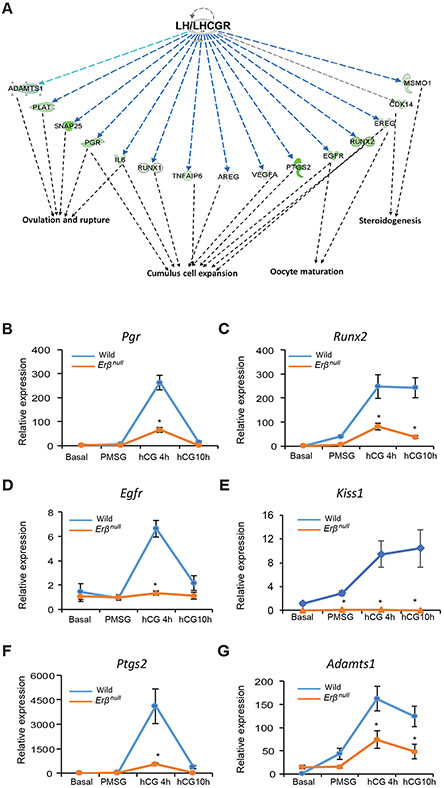
Figure 5. hCG-regulated genes that are dependent on ERβ and involved in follicle maturation.
hCG-regulated genes that were identified in Fig. 3C were subjected to Ingenuity Pathway Analysis (IPA). IPA revealed a subset of LH/LHCGR-regulated genes involved in steroidogenesis, cumulus cell expansion, oocyte maturation, and ovulation (A). The expression of some of the key genes involved in follicle maturation and ovulation were further confirmed by RT-qPCR (B-G). The expression of Pgr, Egfr and Ptgs2 remained unchanged from Basal to PMSG but increased significantly at hCG 4h and then came to basal levels at 10h following hCG in wildtype GCs (B, D, F). The expression levels and pattern of Pgr, Egfr and Ptgs2 in Erβnull GCs were similar to that of wildtype GCs except at hCG4h, where the expression was significantly lower in Erβnull GCs compared to wildtype GCs (B, D, F). The expression of Runx2, Kiss1 and Adamts1 increased slightly from Basal to PMSG but increased rapidly at hCG 4h and then remained unchanged at hCG 10h in Erβnull GCs (C, E, G). The expression pattern of Runx2 and Adamts1 in Erβnull GCs was similar to that of wildtype GCs, but thereafter at hCG4h and hCG 10h expression was significantly lower in Erβnull GCs compared to wildtype GCs (C, G). The expression of Kiss1 remained unchanged from Basal to hCG 10h in Erβnull GCs. In contrast, Kiss1 expression level at PMSG, hCG4h and hCG 10h was significantly higher in wildtype GCs as compared to Erβnull GCs (E). RT-qPCR data are represented as mean ± SEM. n ≥ 6. *P ≤ 0.05.