Figure 3. Activation of Artemis by autophosphorylated DNA-PK on a hairpin-end DNA.
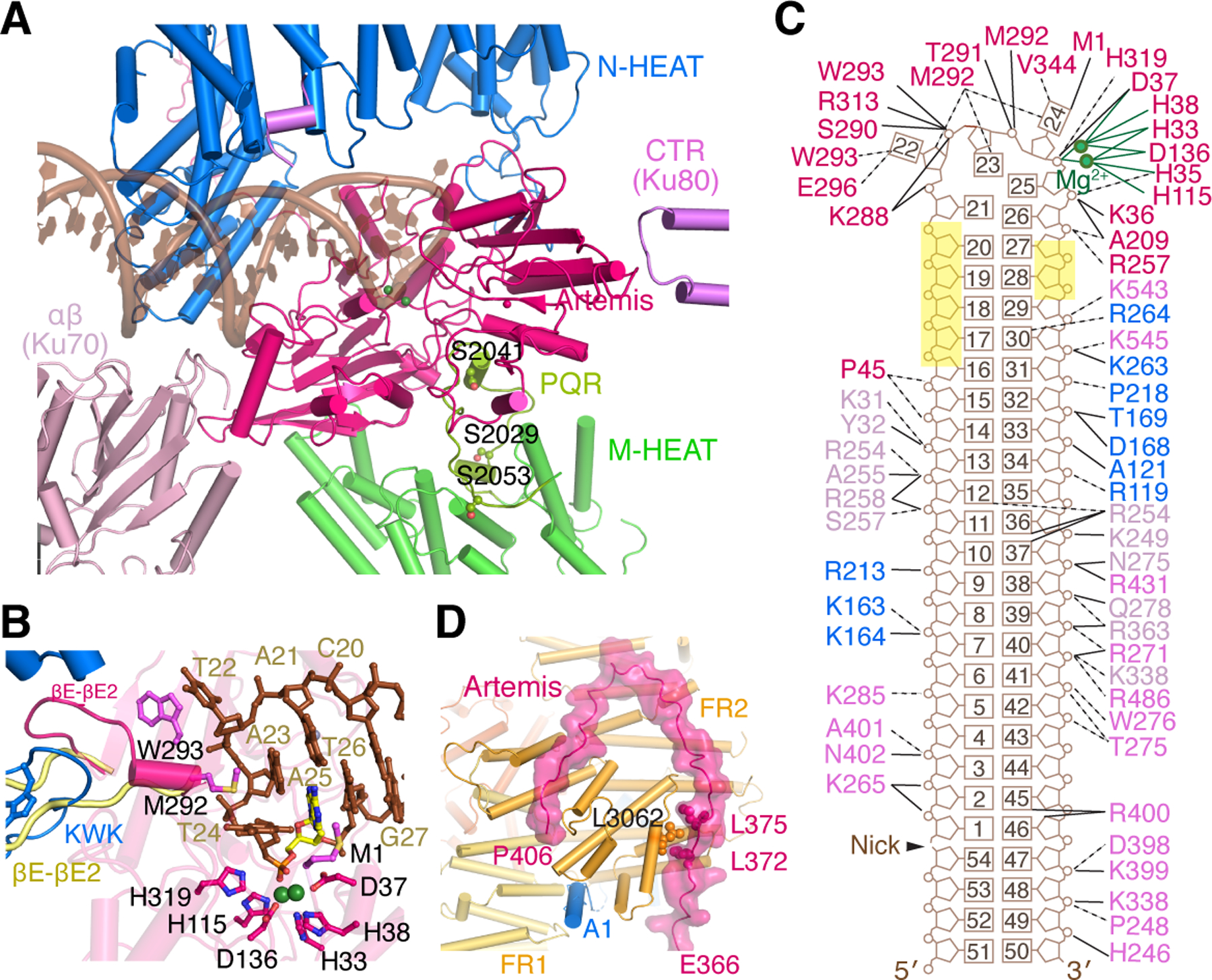
Zoom-in view of interactions between Artemis and DNA-PK with hairpin DNA. The PQR patch (aa 2000–2060), which is unphosphorylated and colored yellow-green, interacts with Artemis on the backside from DNA binding. B. DNA hairpin end in the Artemis active site. The catalytic residues are shown as magenta sticks. Mg2+ ions are shown as green spheres. The nucleotide containing the scissile phosphate is highlighted in yellow. The βE-βE2 loop of apo-Artemis (aa 293–303, shown in pale yellow after structural superposition) is remodeled by the KWK (aa 503–521) and adjacent loop (aa 447–452) of DNA-PKcs. The Artemis βE-βE2 loop forms a helix and contacts the unpaired DNA hairpin end with M292 and W293, which are shown as magenta sticks. C. Diagram of DNA interactions made by Artemis, DNA-PKcs, and Ku. Labels are colored in the same scheme as in panel A. The nucleotides with no protein contact and exposed to solvent are highlighted in yellow. D. Zoom-in view of the extended peptide of Artemis (aa 361–406) reaching to FATKIN of DNA-PKcs, stabilizing the inactive state. Helix A1 of DNA-PKcs (blue) is wedged between FR1 and FR2, further stabilizing the inactive state. L3062 of DNA-PKcs, which is sandwiched by L372 and L375 of Artemis, must be critical for the protein complex formation, as the L3062R mutation leads to SCID.
