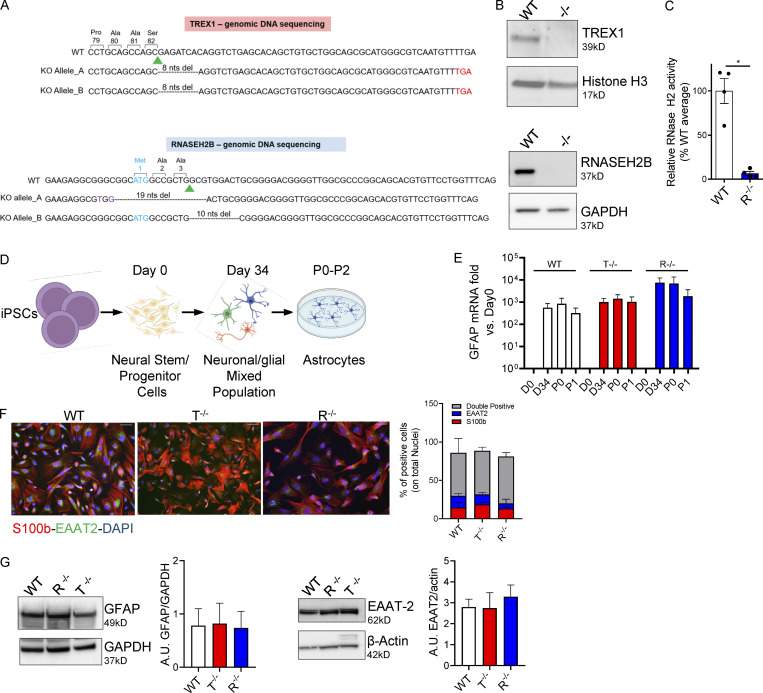
Figure 1.
KO iPSCs efficiently differentiate into proinflammatory astrocytes. (A) Human genomic DNA sequencing. Green arrowhead, sgRNA cut site; purple, nucleotide substitutions; cyan, start codon; red, premature stop codon generated by nonhomologous end joining–mediated out-of-frame repair. Nucleotides deletions are dashed. Reported are the amino acids encoded by the nucleotides upstream of the sgRNA cut site. (B) WB detection of TREX1 and RNASEH2B protein levels in WT and edited clones. (Bottom panel also shown in Fig. S1 A.) (C) RNase H2 enzymatic activity was analyzed in WT and KO iPSCs as cleavage of an in vitro synthetized and labeled RNA/DNA molecule measured by fluorescence. (Mean ± SEM; n = 4 independent differentiations; one-tailed Mann–Whitney U test; *, P < 0.05.) (D) Scheme of proinflammatory astrocytes differentiation protocol created with BioRender.com. (E) GFAP astrocyte marker was detected by gene expression in full KO clones and WT at passage 1 after the iPSC-derived astrocytes enrichment step and expressed as fold vs. day 0, normalized to the HPRT1 housekeeping gene. (Mean ± SEM; n = 5 independent differentiations.) (F) Percentages of S100b- and EAAT2-expressing double-positive cells were measured by IF at passage 1 and quantified. One representative image per genotype is shown. Scale bar, 50 µm. (Mean ± SEM; n = 4 independent differentiations.) (G) Expression and ImageJ quantification of the astrocyte markers EAAT2 and GFAP by WB at passage 1. One representative gel is shown. (Mean ± SEM; n = 3 independent differentiations.) Source data are available for this figure: SourceDataF1.