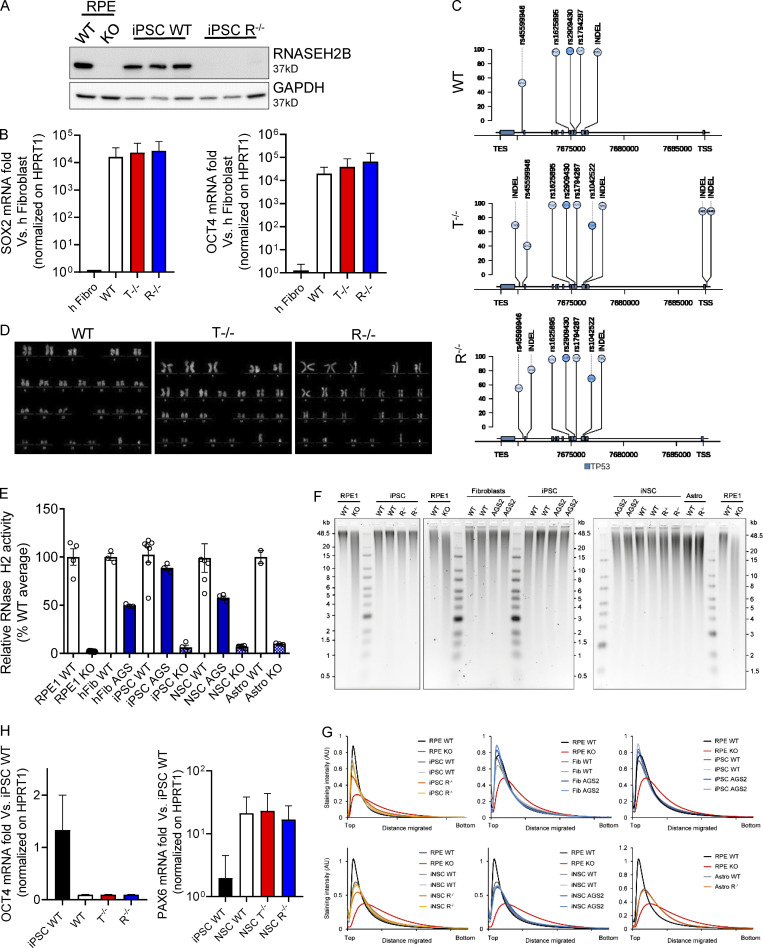
Figure S1.
Characterization of KO iPSCs and iPSC-NSCs. (A) Loss of RNASEH2B expression in RNASEH2BKO iPSCs was confirmed by WB and comparison to RNASEH2BKO hTERT-RPE1 cells. (n = 3 independent experiments; panel also shown in Fig. 1 B.) (B) SOX2 and OCT4 pluripotency markers were detected by gene expression in full KO clones and WT compared with the healthy human fibroblasts (h Fibro) previously used to reprogram these iPSCs and normalized on HPRT1 housekeeping gene. (Mean ± SEM; n = 3 independent experiments.) (C) Lolliplot of TP53 variants in WT, T−/−, and R−/− KO clones. Only single nucleotide polymorphisms (SNPs) and insertion–deletion mutations (INDELs) with a frequency higher than the 20% (see y-axis scale) are reported, and variants located in exonic vs. intronic regions are colored in a different way (light blue and bright blue, respectively). TES, transcription end site. (D) Karyotype of the WT and KO clones. (E) Although cellular RNase H2 activity was substantially reduced in RNASEH2BKO iPSCs, iPSC-NSCs, and astrocytes, as well as in AGS2 (RNASEH2BA177T/A177T) primary fibroblasts, iPSCs and iPSC-NSC KO cells retained low-level nuclease activity against substrate (DNA duplex with a single embedded ribonucleotide), above that seen for independently generated RNase H2–null hTERT-RPE1 cells. Individual data points for n = 2–6 independent differentiations. (Mean ± SEM; n = 2–6 independent experiments.) (F) Representative results for alkaline gel electrophoresis of RNase H2–digested genomic DNA from WT, RNASEH2BKO (R−/−), and RNASEH2BA177T/A177T (AGS2) cells (primary fibroblasts, iPSCs, iPSC-NSCs, and astrocytes). hTERT-RPE1 RNase H2–null (RPE1 KO) DNA was included as a positive control for increased fragmentation as a consequence of embedded ribonucleotides. (n = 2–3 technical replicates.) (G) Densitometry plots for gels in A show no apparent shift in DNA fragment size distribution for mutant RNASEH2B fibroblasts, iPSCs, iPSC-NSCs, or astrocytes, in contrast with independent RNase H2–null RPE1 cells. (H) OCT4 and PAX6 were detected by gene expression in differentiated WT and KO clones in comparison with WT iPSCs and normalized on HPRT1 housekeeping gene. (Mean ± SEM; n = 3 independent experiments.) Source data are available for this figure: SourceDataFS1.