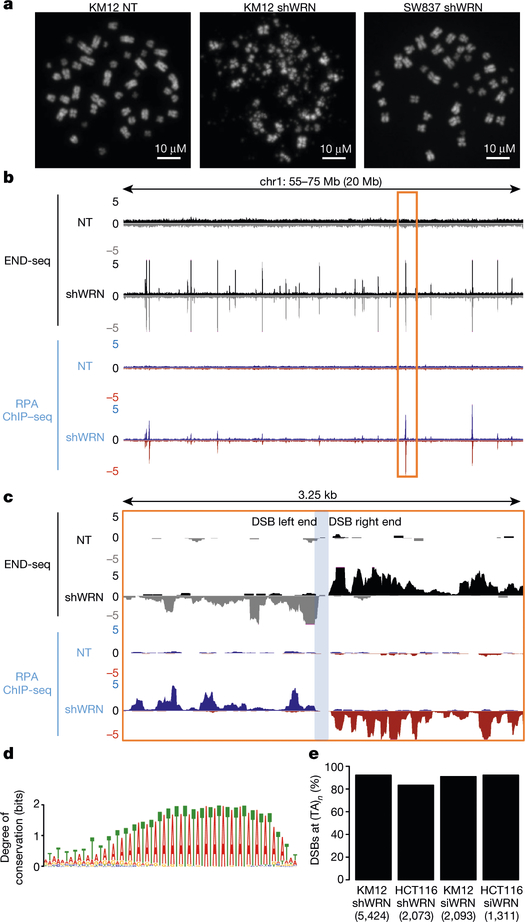
Fig. 1 |. WRN depletion in MSI cells induces recurrent DSBs at (TA)n dinucleotide repeats.
a, Representative metaphase spreads from KM12 and SW837 cells containing an inducible WRN shRNA (shWRN) cassette. Cells were treated with DMSO (NT) or doxycycline (shWRN) for 48 h. Data are representative of three independent experiments, n = 100 metaphases for each condition. b, Genome browser screenshots displaying END-seq (n = 5) and RPA-bound ssDNA ChIP–seq (n = 1) profiles as normalized read density (reads per million, RPM) for KM12-shWRN cells. Positive- and negative-strand END-seq reads are displayed in black and grey, and positive- and negative-strand RPA–ssDNA ChIP–seq reads are in blue and red, respectively. c, Genome browser screenshot zoomed in on the highlighted region in b (orange box). The light-blue shading indicates the gap region between the left and right ends of the DSB. d, Motif analysis for sequence enrichment in the gap between positive and negative END-seq peaks in KM12-shWRN cells. e, Fraction of END-seq peaks occurring at sites of (TA)n repeats for KM12-shWRN and HCT116-shWRN cells treated with either doxycycline (shWRN, n = 2 for HCT116) or WRN siRNAs (siWRN, n = 1) for 72 h. Numbers in parentheses indicate number of peaks identified by peak-calling.