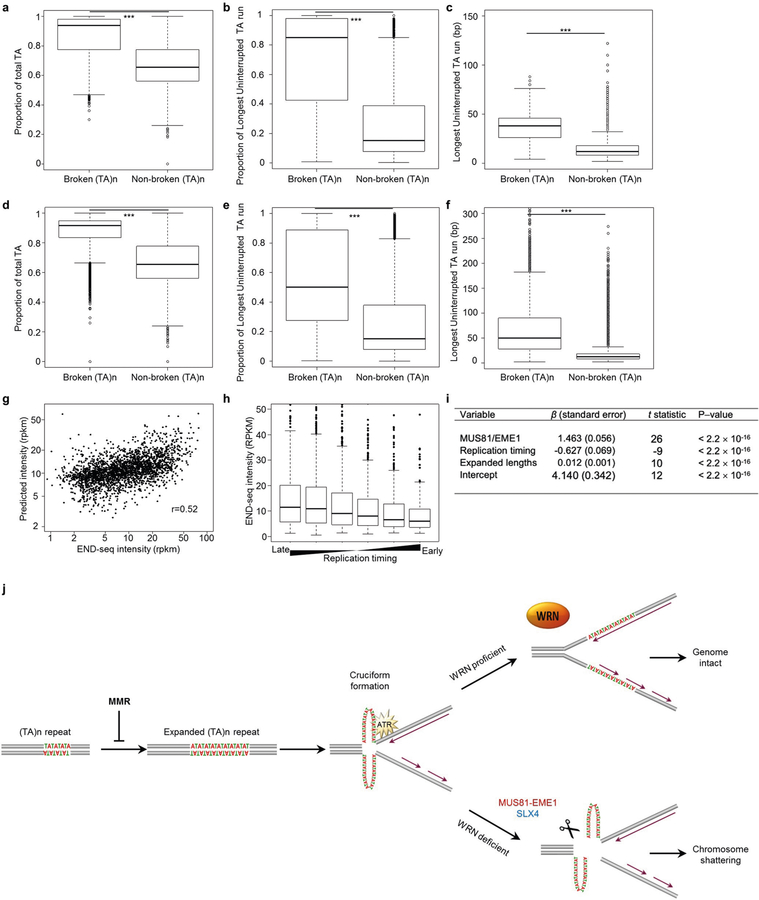
Extended Data Fig. 8 |. Large-scale expansions occur at long, uninterrupted (TA)n repeat sequences.
(a) Boxplot showing, in the hg19 reference genome, the proportion of (TA)n repeat units found within the full annotated sequence at broken or non-broken (TA)n repeats in KM12 cells. n = 5,400 (broken) and n = 59,729 (non-broken) sites were examined for statistical significance using one-sided Wilcoxon rank sum test. ***P < 2.2 × 10−16. b, Box plot showing, in the hg19 reference genome, the proportion of the longest run of uninterrupted (TA)n within the full annotated sequence at broken or non-broken (TA)n repeats in KM12 cells. n = 5,400 (broken) and n = 59,729 (non-broken) sites were examined for statistical significance using one-sided Wilcoxon rank sum test. ***P < 2.2 × 10−16. c, Box plot showing, in the hg19 reference genome, the length (bp) of the longest uninterrupted (TA)n dinucleotide repeats within the full annotated sequence at broken or non-broken (TA)n repeats in KM12 cells. n = 5,400 (broken) and n = 59,729 (non-broken) sites were examined for statistical significance using one-sided Wilcoxon rank sum test. ***P < 2.2 × 10−16. d, Box plot showing, in long read sequencing data, the proportion of (TA)n repeat units found within the full sequence at broken or non-broken (TA)n repeats in KM12 cells. n = 5,400 (broken) and n = 61,244 (non-broken) sites were examined for statistical significance using one-sided Wilcoxon rank sum test. ***P < 2.2 × 10−16. e, Box plot showing, in long-read sequencing data, the proportion of the longest run of uninterrupted (TA)n within the full sequence at broken or non-broken (TA)n repeats in KM12 cells. n = 5,400 (broken) and n = 61,244 (non-broken) sites were examined for statistical significance using one-sided Wilcoxon rank sum test. ***P < 2.2 × 10−16. f, Boxplot showing, in long-read sequencing data, the length (bp) of the longest uninterrupted (TA)n dinucleotide repeat within the full sequence at broken or non-broken (TA)n repeats in KM12 cells. n = 5,400 (broken) and n = 61,244 (non-broken) sites were examined for statistical significance using one-sided Wilcoxon rank sum test. ***P < 2.2 × 10−16. g, Multiple linear regression model predicting END-seq peak intensity of KM12-shWRN cells treated with doxycycline (shWRN) for 72 h derived from END-seq intensity of MUS81–EME1 cleavage in situ, replication timing, and expanded length of broken (TA)n. The Pearson correlation coefficient is indicated (see i). h, END-seq intensity of broken (TA)n repeats in KM12-shWRN cells treated with doxycycline for 72 h grouped by replication timing values from late replicating to early replicating. i, Multiple linear regression was performed to predict END-seq peak intensity of KM12-shWRN cells treated with doxycycline for 72 h based on following parameters: END-seq intensity of MUS81–EME1 cleavage in situ, replication timing, and expanded length of broken (TA)n. END-seq intensity upon shWRN induction and MUS81–EME1 cleavage were calculated using RPKM in ±1 kb window around broken (TA)n. Mean value was used for replication timing quantification. Expanded lengths were identified from long read sequencing data. Estimates of the standardized regression coefficients (β) are shown, along with t-statistics and P values based on the standardized coefficients. j, Model for MSI cell dependence on WRN. Large-scale expansions of (TA)n repeats are associated with MSI in MMR-deficient cells. When (TA)n reach above a critical length, they extrude into cruciform-like structures, which stall replication forks and activate ATR kinase, which in turn phosphorylates WRN and other substrates to complete DNA replication. In the absence of WRN, MUS81–EME1 or SLX4 cleaves secondary structures at (TA)n repeats, thereby shattering the chromosomes. All box plots are as in Fig. 2a, b.