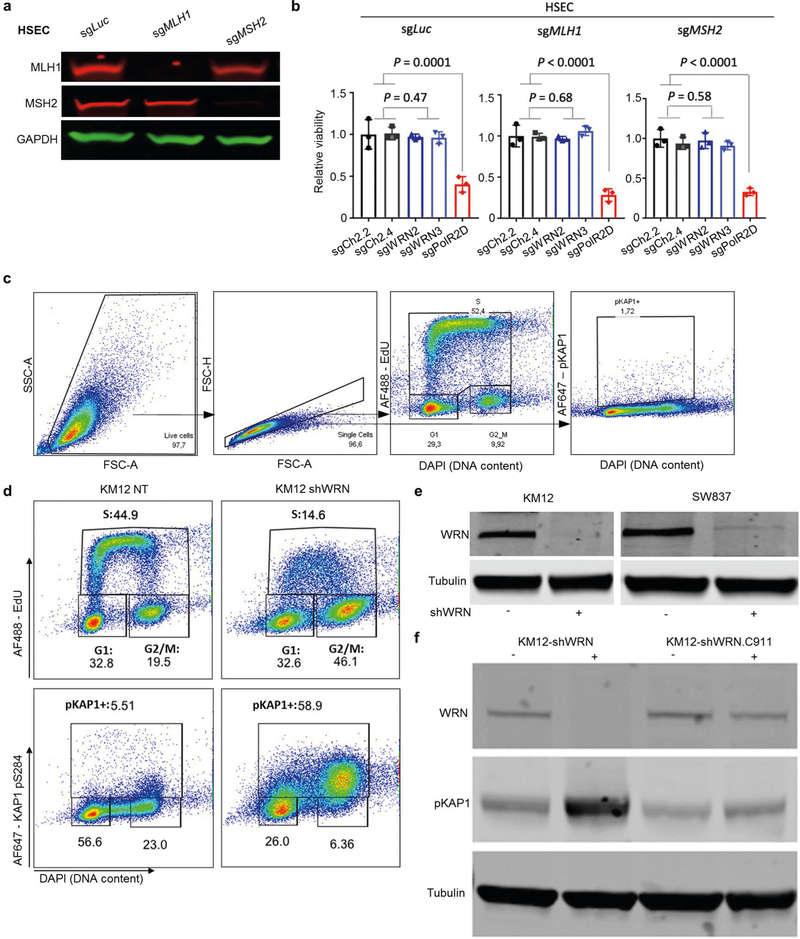
Extended Data Fig. 1 |. WRN depletion induces DNA damage in different MSI cell lines.
a, Western blot analysis of MLH1, MSH2, and GAPDH protein levels in HSECs after CRISPR–Cas9 knockout. sgLuc, control single-guide RNA (sgRNA) targeting luciferase; sgMLH1 and sgMSH2, sgRNAs targeting MLH1 and MSH2, respectively. For gel source data, see Supplementary Fig. 1. b, Relative viability 7 days after sgRNA transduction in HSECs. sgCh2.2 and sgCh2.4 denote negative controls targeting chromosome 2 intergenic sites; sgPolR2D denotes a pan-essential control. sgWRN2 and sgWRN3 denote experimental sgRNA targeting WRN. Data are mean and s.d. P values were determined using two-tailed Student’s t-test (n = 3). c, Example of flow cytometry gating strategy used in d and Extended Data Fig. 4c. d, Flow cytometry profiles for exponentially growing KM12-shWRN cells treated with DMSO (NT) or doxycycline (shWRN) for 72 h. EdU was added during the last 30 min before collecting cells. Percentage of cells in the gates is indicated. Data are representative of three independent experiments. e, Western blot analysis of WRN protein levels in KM12-shWRN and SW837-shWRN treated with DMSO or doxycycline for 72 h. Data are representative of three independent experiments. For gel source data, see Supplementary Fig. 1. f, Western blot analysis of WRN and pKAP1 protein levels in KM12-shWRN and KM12-shWRN. C911 (non-targeting shRNA) treated with DMSO or doxycycline for 72 h. Data are representative of three independent experiments. For gel source data, see Supplementary Fig. 1.