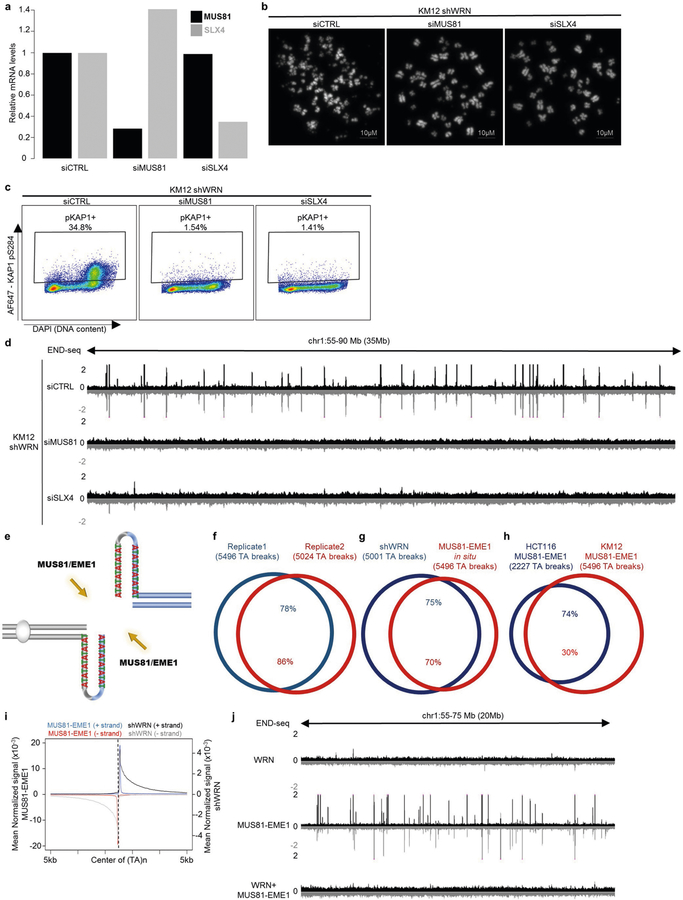
Extended Data Fig. 4 |. (TA)n repeat-forming repeats in MSI cell lines are substrates for MUS81–EME1.
a, Quantitative PCR with reverse transcription (qRT–PCR) analysis quantification (n = 1) of MUS81 and SLX4 mRNA levels in KM12-shWRN cells transfected with non-targeting siRNAs (siCTRL), MUS81 siRNAs (siMUS81), or SLX4 siRNAs (siSLX4). b, Representative images of metaphase spreads from KM12-shWRN cells treated with doxycycline (shWRN) and non-targeting siRNAs (siCTRL), MUS81 siRNAs (siMUS81), or SLX4 siRNAs (siSLX4) for 48 h. Data are representative of three independent experiments, n = 100 metaphases for each condition. c, Flow cytometric profiles for KAP1 phosphorylation in exponentially growing KM12-shWRN cells treated with doxycycline (shWRN), plus non-targeting siRNAs (siCTRL), MUS81 siRNAs (siMUS81), or SLX4 siRNAs (siSLX4) for 72 h. Data are representative of three independent experiments. d, Genome browser screenshot displaying END-seq profiles as normalized read density (RPM) for KM12-shWRN cells treated with doxycycline (shWRN), plus non-targeting siRNAs (siCTRL), MUS81 siRNAs (siMUS81), or SLX4 siRNAs (siSLX4) for 72 h. e, Schematic representation of DNA cruciform cleavage by MUS81–EME1 structure-specific endonuclease. f, Venn diagram displaying overlap of END-seq TA breaks between two biological replicates of DMSO-treated KM12-shWRN cells processed with purified recombinant MUS81–EME1 enzyme in situ (MUS81–EME1). n = 1,000 random datasets were generated to test significance of overlap using one-sided Fisher’s exact test (P < 2.2 × 10−16). g, Venn diagram showing overlap in TA breaks between KM12-shWRN cells treated with doxycycline (shWRN) for 72 h, and DMSO-treated cells processed with MUS81–EME1 enzyme in situ (MUS81–EME1). n = 1,000 random datasets were generated to test significance of overlap using one-sided Fisher’s exact test (P < 2.2 × 10−16). h, Venn diagram displaying overlap between TA breaks from KM12-shWRN and HCT116-shWRN genomic DNA processed in situ with MUS81–EME1 in situ (n = 1 for HCT116). n = 1,000 random datasets were generated to test significance of overlap using one-sided Fisher’s exact test (P < 2.2 × 10−16). i, Genome-wide aggregate analysis of END-seq signal around TA breaks from KM12-shWRN cells treated with doxycycline for 72 h (shWRN) (black denotes positive-strand reads, grey denotes negative-strand reads), or DMSO-treated KM12-shWRN cells processed with purified recombinant MUS81–EME1 enzyme in situ (blue denotes positive-strand reads, red denotes negative-strand reads). j, Genome browser screenshot displaying END-seq profiles for DMSO-treated KM12-shWRN cells (WRN proficient) processed in situ with either purified recombinant WRN, MUS81–EME1, or WRN followed by MUS81–EME1. For the latter, proteinase K digestion was performed between the two enzymatic treatments.