Figure 1, Key Figure. The main polygenic component of malaria resistance is not enriched in African populations.
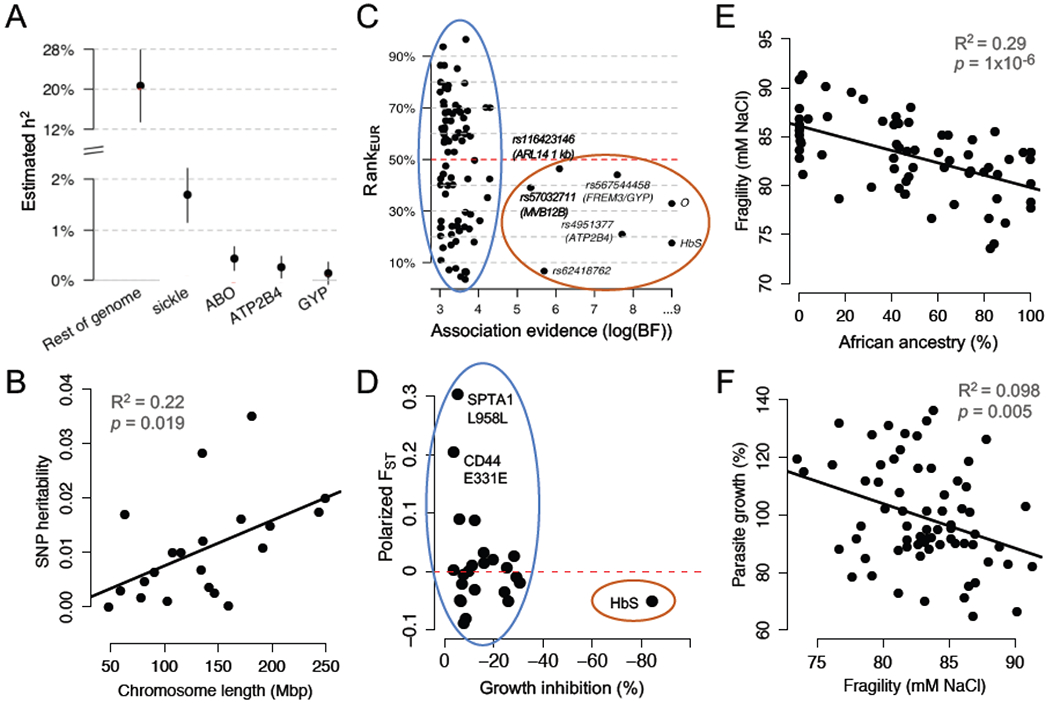
(A) 89% of the estimated heritability of severe malaria risk is attributed to variants with effects too small to be confidently detected by GWAS. Reproduced from [7]. (B) SNP-heritability of severe malaria risk is distributed across chromosomes proportional to their length. Replotted from [36]. (C) Strongly associated (large-effect) alleles from a GWAS for severe malaria risk occur at higher frequencies in African populations, compared to European populations (red oval). Smaller-effect alleles, which are less confidently associated, are equally likely to be more frequent in European populations (blue oval). RankEUR is the conditional rank of the allele in European populations, given African allele count; values greater than/less than 50% indicate that the allele is more common in Europe/Africa. Reproduced from [7]. (D) Alleles from healthy RBCs associated with reduced P. falciparum growth rate in vitro are not enriched for higher frequencies in African populations (blue oval). Polarized FST indicates the differentiation in allele frequencies between African and European populations in 1000 Genomes; values greater than/less than 0 indicate that the allele is more common in Europe/Africa. Replotted from [15]. (E) In donors who lack large-effect alleles for malaria resistance, red cell osmotic fragility is negatively related to exome-wide African ancestry. Replotted from [15]. (F) In donors who lack large-effect alleles for malaria resistance, higher red cell osmotic fragility predicts slower P. falciparum growth in vitro. Replotted from [15]. Panels A and C are reproduced with minor modifications under a Creative Commons license (https://creativecommons.org/licenses/by/4.0/).
