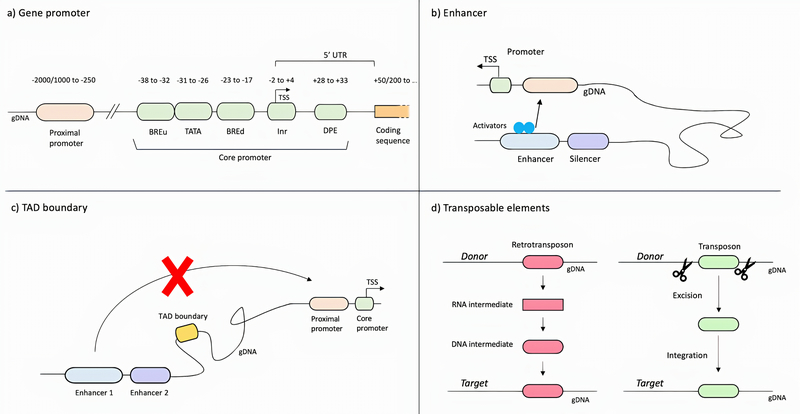
Figure 2:
Schematic representation of non-coding regulatory elements. a) Gene promoters represent the site where the transcriptional machinery assembly occurs, and transcription factors interact to regulate the expression of genes. Gene promoters include the core promoter, the minimal required portion to initiate the transcription, and the proximal promoter. b) Enhancers are regulatory elements that can be bound by activators and repressor and modulate the expression of target genes. c) TAD boundaries may prevent the physical interaction between enhancers and the target genes. d) Transposable elements are regulatory sequences capable of altering their position in the genome. Retrotransposons use a “copy and paste” mechanism and introduce a copy of themselves to a different genetic location; transposons use a “cut and paste” mechanism: they excise themselves from one genetic locus and integrate into a different one. Translocation of TE occurs predominantly in germ cells and during early embryogenesis, when TEs contribute defining the temporal expression of genes. BREu: upstream TFIIB Recognition Element, BREd: downstream TFIIB Recognition Element, TATA box, Inr: Initiator element, DPE: downstream promoter element, TSS: transcription start site, gDNA: genomic DNA.