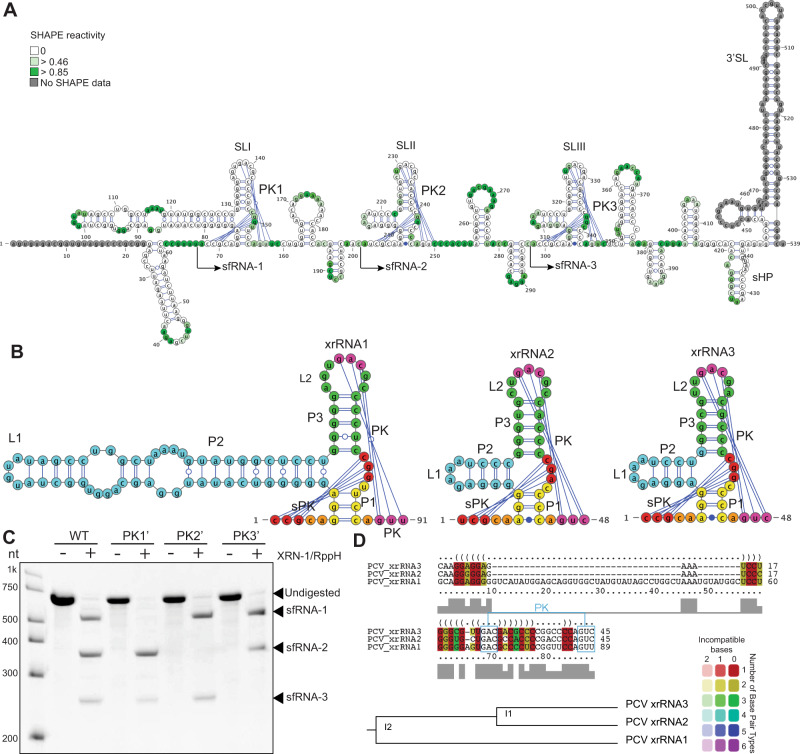
Fig. 4. cISFs PCV contains multiple copies of divergent class 1b xrRNAs.
A Secondary structure of PCV 3’UTR generated using SHAPE-assisted folding. Shading intensity indicates NMIA reactivity. Values are the means from 3 independent experiments, each with technical triplicates. SL – stem loop, PK – pseudoknot. B Secondary structure of PCV xrRNAs. Each element of the secondary structure is shown in colour. C In vitro XRN1 resistance assay with WT and mutated PCV 3’UTRs. PK1’, PK2’ and PK3’ mutations were introduced into the terminal loop regions of SLI, SLII and SLIII, respectively, and represented the GAC - > CUG change in the PK-forming region. Image is representative from three independent experiments that showed similar results. D Alignment of sequence and structure of PCV xrRNAs was performed using LocARNA.