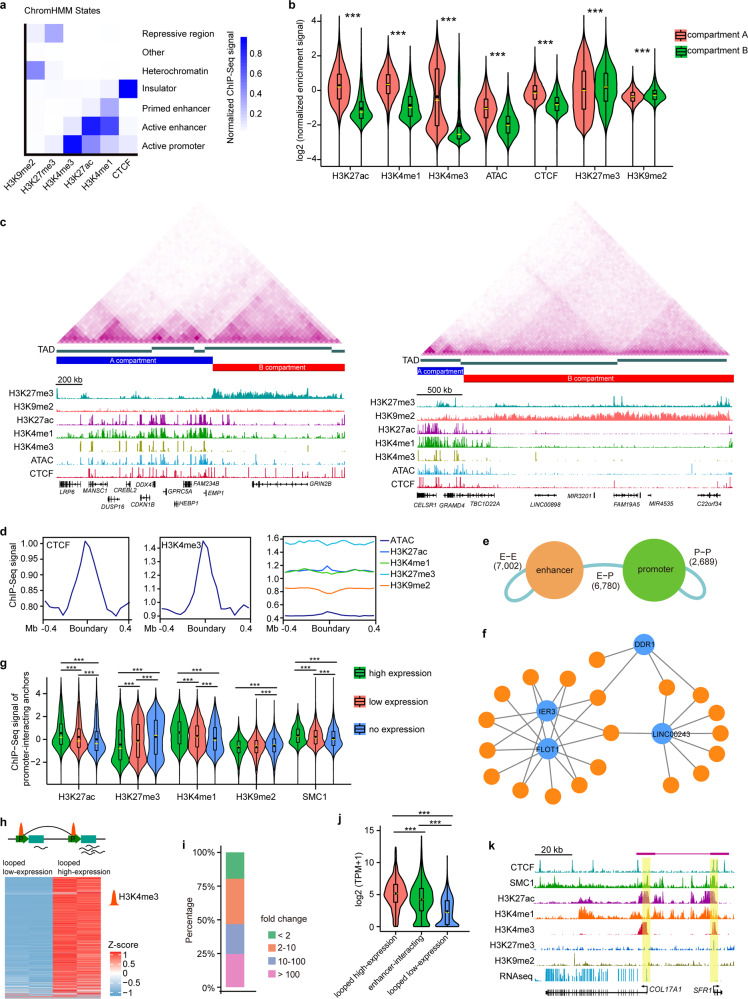
Fig. 3. Characterization of a 3D epigenetic regulatory map in LSCs.
a ChromHMM state annotation using the indicated ChIP-Seq data. b Violin and boxplots showing normalized enrichment signal of ATAC-Seq and the indicated ChIP-Seq data at A (n = 12,822) and B (n = 13,324) compartments. ***p < 0.001 from two-way ANOVA. c chromatin interaction heatmaps, identified TADs, chromatin compartments and tracks for enrichment of ATAC-Seq and the indicated ChIP-Seq data at the denoted genomic loci. d Metaplots showing the enrichment signal of ATAC-Seq and the indicated ChIP-Seq data at the TAD boundaries. e Numbers of E–E, E–P, and P–P loops. f Selected E–P interaction network. Orange nodes represent enhancers and blue nodes represent promoters. Edges represent chromatin interactions. g Violin and boxplots showing normalized enrichment signal for the indicated ChIP-Seq data at the anchors connected to gene promoters with high (n = 8429), low (n = 7288) and no (n = 7316) expression. High-expression genes: TPM ≥ 10; Low-expression genes: 10 > TPM ≥ 0.1; No expression genes: TPM < 0.1. ***p < 0.001 from two-way ANOVA. h Gene expression heatmap of interaction pairs between looped low-expression and looped high-expression promoters. i Percentages of the indicated fold changes of looped high-expression promoters/looped low-expression promoters. j Violin and boxplots showing gene expression levels of looped high-expression (n = 1122), enhancer-interacting (n = 2230) and looped low-expression (n = 1126) promoters. ***p < 0.001 from two-way ANOVA. k A identified chromatin loop and tracks for RNA-Seq and the indicated ChIP-Seq enrichment at the COL17A1 and SFR1 loci. All the boxplots indicate the 25th percentile (bottom of box), median (horizontal yellow line inside box), mean value (dark spot inside box), and 75th percentile (top of box). Whiskers indicate 1.5 times the interquartile range.