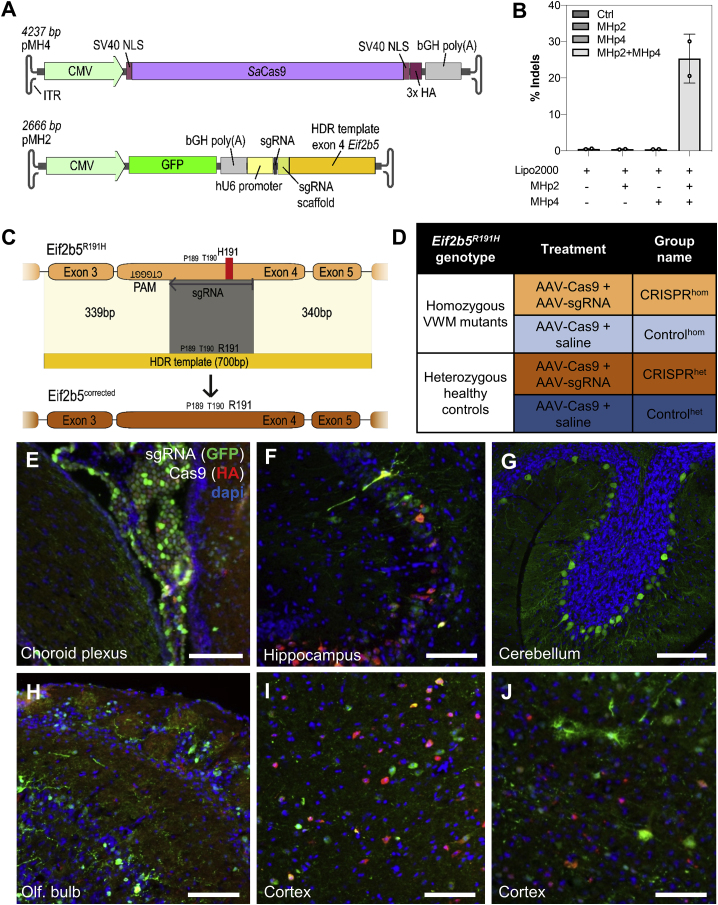
Figure 1.
Overview of the CRISPR/Cas9 system, experimental setup, and transduction patterns in vivo
(A) Schematic overview of the dual AAV constructs. (B) In vitro testing of the sgRNA. HEK293T reporter cells were transfected with the respective treatment using Lipofectamine 2000 (Lipo2000). Insertions and deletion (indel) frequency was determined by next-generation targeted amplicon sequencing. Values represent mean ± SD of two technical replicates. (C) Graphical illustration of the used HDR design. The Eif2b5R191H disease-causing mutation (red) on exon 4 was targeted by a 21 bp long sgRNA (gray) to induce a double-stranded DNA (dsDNA) break. The disease-causing mutation lies in the center of the 700 bp long HDR template (orange). The HDR template with homology-arms of 349 bp and 350 bp corrects the R191H mutation and additionally introduces a silent mutation in the sgRNA seed region. (D) Overview of treatment groups for in vivo studies (n = 79 per group). Cells expressing Cas9 (tagged with HA, red) and/or sgRNA and DNA donor template (tagged with GFP, green) in (E) the choroid plexus, (F) hippocampus, (G) cerebellum, (H) olfactory bulb, (I) deeper cortical layers, and (J) superficial layers of the cortex of animals between 5 and 9 weeks of age. Scale bars represent 75 μm.