Fig. 3.
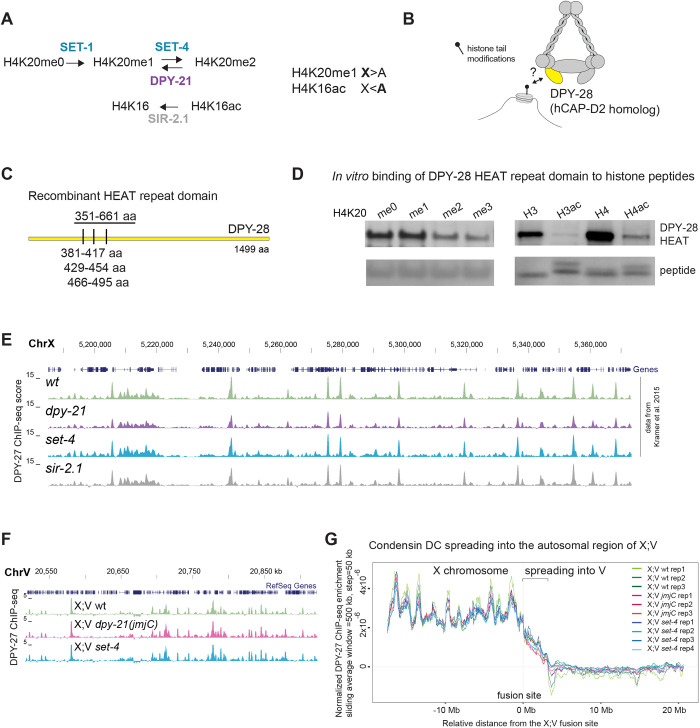
Condensin DC may interact with histone tails, but set-4, sir-2.1 and catalytic activity of DPY-21 do not regulate condensin DC binding as measured by ChIP-seq. (A) Enzymes that regulate H4K20 methylation and H4K16 acetylation. In hermaphrodites, H4K20me1 is increased and H4K16ac is reduced on the dosage compensated X chromosomes (X) compared to autosomes (A). The dpy-21 null mutant is the (e418) allele with a premature stop codon that eliminates the protein (Yonker and Meyer, 2003), the dpy-21(JmjC) mutant is the (y607) allele, a point mutation that nearly abolishes H4K20me2 demethylase activity without eliminating the protein itself (Brejc et al., 2017). The set-4 null mutant is (n4600), a knockout allele that eliminates H4K20me2 and H4K20me3 (Delaney et al., 2017). The sir-2.1 null mutant is (ok434), a knockout allele that increases H4K16ac (Wells et al., 2012). (B) Cartoon depicting possible interaction of HEAT repeat-containing domain of DPY-28 (homologous to human hCAP-D2) with histone tail modifications. (C) Three HEAT repeats annotated by Pfam are shown as tick marks. The amino acids (aa) 351–661 were purified and used in peptide binding assays. (D) In-solution peptide binding assay was performed using GST-tagged DPY-28 HEAT domain and biotinylated histone N-terminal tail peptides with the indicated modifications (H3ac and H4ac indicate tetra-acetylated histone H3 and H4 peptides, respectively). The recombinant protein was incubated with peptides bound to magnetic streptavidin beads, and bound fractions were analyzed using western blot. The streptavidin signal below indicates the amount of peptide in each fraction. Methyl modified histone peptide blots representative of two replicates; acetyl and unmodified histone peptides representative of two replicates. (E) UCSC genome browser (https://genome.ucsc.edu/) shot of a representative region of the X chromosome (ChrX) showing similar DPY-27 ChIP-seq patterns in the sir-2.1 null mutant. Data from wild-type N2, the dpy-21 null mutant and the set-4 null mutant are from Kramer et al. (2015) and are plotted for comparison. Chromosome locations are marked in kb. (F) Genome browser view of DPY-27 ChIP-seq enrichment across the fusion site on the autosomal region of the X;V chromosome in X;V wild-type, dpy-21(JmjC) and set-4 null backgrounds. (G) A moving average of the DPY-27 ChIP enrichment score is plotted with a window size of 500 kb and step size of 50 kb in X;V fusion strains with wild-type, dpy-21(JmjC) and set-4 null backgrounds. DPY-27 ChIP-seq data was normalized to reduce variability between replicates by z-score standardization of ChIP/input ratios to the background from autosomes I–IV, followed by equalization of total ChIP-seq signal to 1 in X;V.